The SPARC complex defines RNAPII promoters in Trypanosoma brucei
Figures
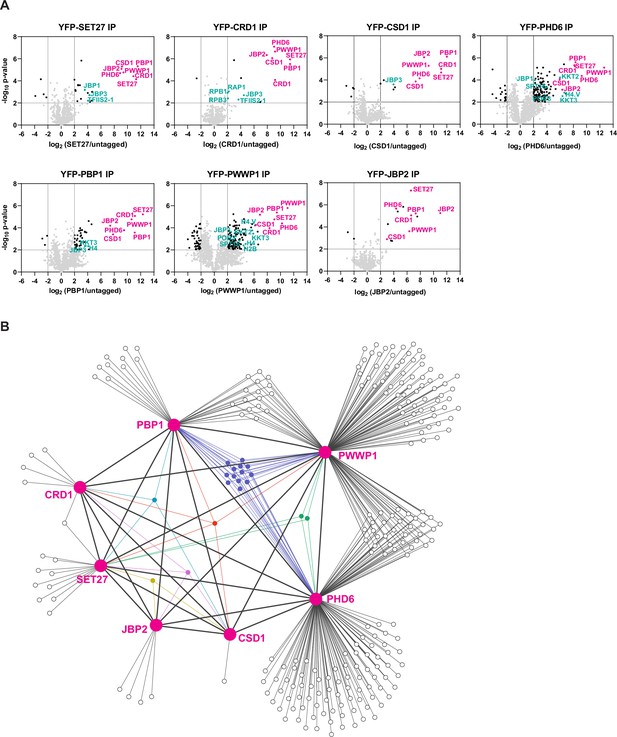
Identification of SPARC in bloodstream form Trypanosoma brucei.
(A) Proteins previously shown to be enriched in CRD1 and SET27 coimmunoprecipitations (co-IPs) were YPF-tagged and analyzed by affinity selection and LC-MS/MS to identify their protein interaction partners. Volcano plots are based on three biological replicates for each sample. Cutoffs used for significance: log2 (tagged/untagged)>2 or <−2 and Student’s t-test p-value<0.01. See Supplementary file 1 for a complete list of proteins enriched in each affinity selection. Putative SPARC complex subunits are shown in pink and other proteins of interest are shown in teal. The CRD1 and SET27 co-IPs are reproduced from Figure 4A by Staneva et al., 2021. (B) Interaction network of the proteins enriched in the co-IP experiments shown in (A). SPARC components are connected with thick lines while all other interactions are shown with thin lines. Proteins which interact with three or more SPARC components are represented in different colors. See Supplementary file 2 for a complete list of shared and unique interactors in these co-IPs.
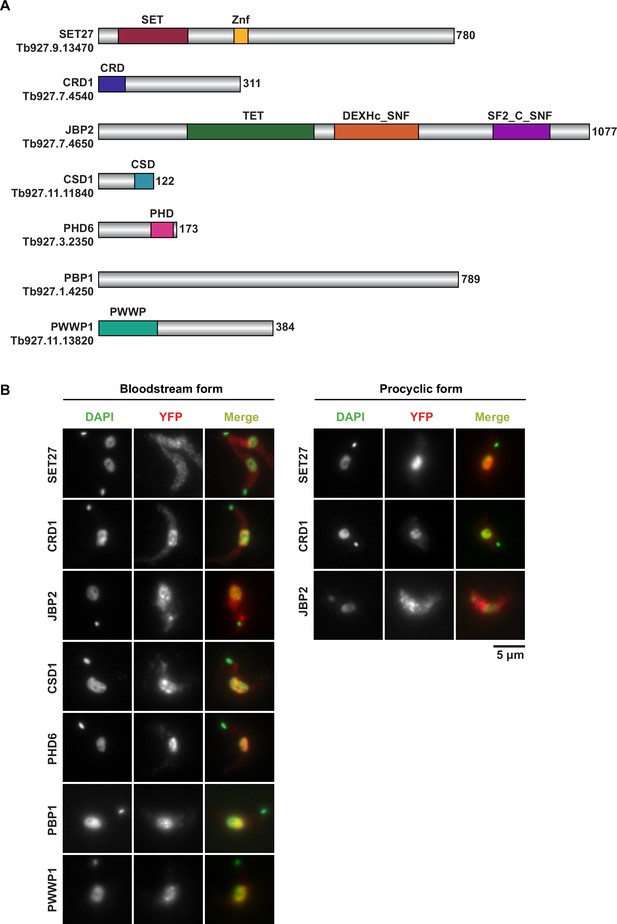
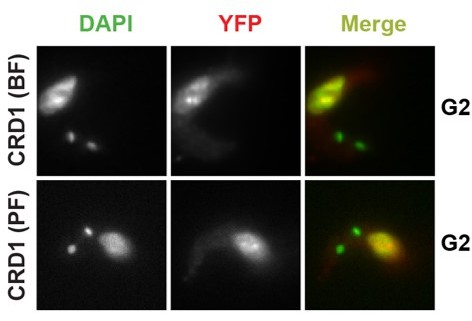
Domain architecture and localization of SPARC complex subunits.
(A) Conserved domains and sequences of core SPARC complex subunits and JBP2. (B) Immunolocalization of YFP-tagged core SPARC components and JBP2 in bloodstream and procyclic form cells. In merge panels, YFP-tagged proteins are shown in red and DAPI-stained DNA is in green. SET27 (BF) image panels are reproduced from Figure 1 by Staneva et al., 2021.
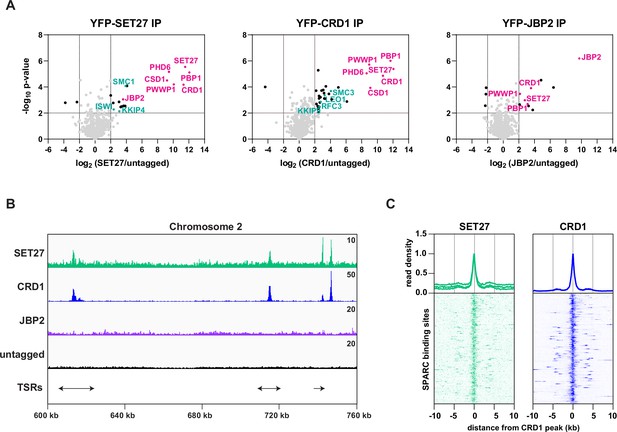
SPARC is present in procyclic form cells.
(A) SET27, CRD1, and JBP2 were YPF-tagged in PF cells and analyzed by affinity selection and LC-MS/MS to identify their protein interaction partners. Volcano plots are based on three biological replicates for each sample. Cutoffs used for significance: log2 (tagged/untagged)>2 or <−2 and Student’s t-test p-value<0.01. See Supplementary file 1 for a complete list of proteins enriched in each affinity selection. Core SPARC components and the auxiliary subunit JBP2 are shown in pink and other proteins of interest are shown in teal. (B) A region of Chromosome 2 (same as in Figure 2A) is shown with ChIP-seq reads mapped for the indicated proteins. A single replicate is shown for each protein. ChIP-seq performed in cells lacking any tagged protein (untagged) was included as a negative control. Tracks are scaled separately as fragments per million (the exact value is indicated in the top-right corner of each track). The positions of single and double transcription start regions (sTSRs and dTSRs) are shown below with arrows indicating the direction of transcription. (C) Enrichment profiles for SPARC components. CRD1 is used as a reference because it has the most prominent peaks at TSRs. The metagene plots (top) show normalized read density around all CRD1 peak summits, with each replicate plotted separately. The heatmaps (bottom) show protein density around 177 individual CRD1 peaks. Each heatmap shows the average of at least two replicates.
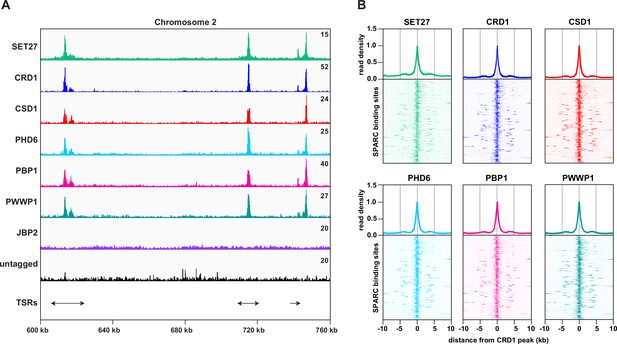
SPARC components target the same genomic loci in bloodstream form Trypanosoma brucei.
(A) A region of Chromosome 2 is shown with ChIP-seq reads mapped for the indicated proteins. A single replicate is shown for each protein. ChIP-seq performed in wild-type cells lacking any tagged protein (untagged) was included as a negative control. Tracks are scaled separately as fragments per million (the exact value is indicated in the top-right corner of each track). The positions of single and double transcription start regions (sTSRs and dTSRs) are shown below with arrows indicating the direction of transcription. ChIP-seq data for CRD1, SET27, and the untagged parental cell line are reproduced from Figure 2A by Staneva et al., 2021. (B) Enrichment profiles for SPARC components. CRD1 is used as a reference because it has the most prominent peaks at TSRs. The metagene plots (top) show normalized read density around all CRD1 peak summits, with each replicate plotted separately. The heatmaps (bottom) show protein density around 177 individual CRD1 peaks. Each heatmap shows the average of at least two replicates. CRD1 and SET27 metagene plots and heatmaps are reproduced from Figure 2C by Staneva et al., 2021.
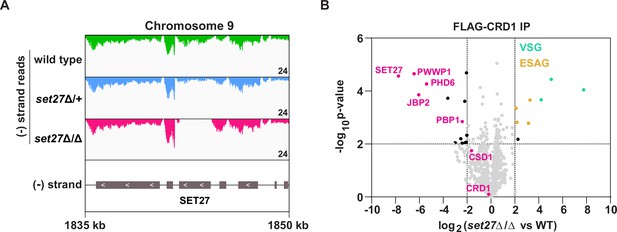
SPARC integrity is compromised in bloodstream form T. brucei lacking SET27.
(A) Tracks showing the distribution of RNA-seq reads in the vicinity of the SET27 gene in wild-type, set27Δ/+and set27Δ/Δ cells. All tracks are scaled identically (number of reads shown in the bottom right corner of each track). (B) Affinity selection of FLAG-CRD1 from wild-type and set27Δ/Δ cells. The volcano plot is based on three biological replicates for each sample. Cutoffs used for significance: log2 (set27Δ/Δ vs. WT)>2 or <−2 and Student’s t-test p-value<0.01. See Supplementary file 1 for a complete list of proteins in these affinity selections. SPARC components are shown in pink, VSGs in green, and ESAGs in yellow.
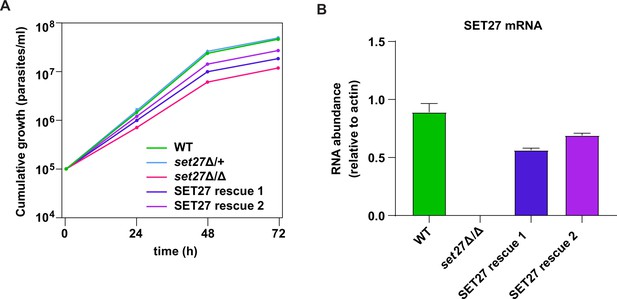
Validation of the SET27 knockout.
(A) Growth curves for wild-type, set27Δ/+, set27Δ/Δ, and the two SET27 rescue clones. Error bars: standard deviation (SD) of three biological replicates. Cells lacking the SET27 gene (set27Δ/Δ) grew substantially slower than WT or set27Δ/+ cells (t-test p-value<0.0001). SET27 rescue clones 1 and 2 also grew significantly slower than WT or set27Δ/+ cells (t- test p-value<0.01) but faster than set27Δ/Δ cells (t-test p-value<0.01). Average doubling times: 6.2 hr (wild-type), 6.0 hr (set27Δ/+), 8.3 hr (set27Δ/Δ), 7.4 hr (SET27 rescue clone 1), and 6.8 hr (SET27 rescue clone 2). (B) SET27 mRNA levels assessed by RT-qPCR in wild-type, set27Δ/Δ, and the two SET27 rescue clones. Error bars: SD of two biological replicates.
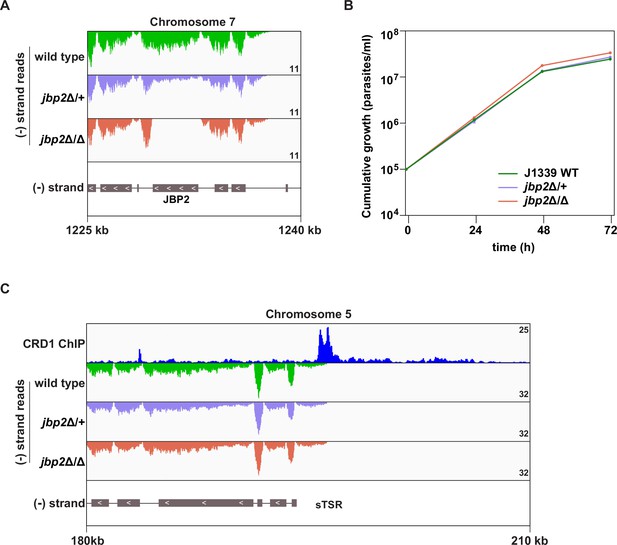
Validation of the JBP2 knockout.
(A) Tracks showing the distribution of RNA-seq reads in the vicinity of the JBP2 gene in wild-type, jbp2Δ/+, and jbp2Δ/Δ cells. All tracks are scaled identically (number of reads shown in the bottom right corner of each track). (B) Growth curves for J1339 wild-type, jbp2Δ/+, and jbp2Δ/Δ cell lines. Error bars: standard deviation (SD) of three biological replicates. Cells lacking the JBP2 gene (jbp2Δ/Δ) grew slightly faster than WT cells (t-test p-value<0.05) and about the same as jbp2Δ/+ cells (t-test p-value=0.05). Average doubling times: 6.9 hr (J1339 WT), 6.8 hr (jbp2Δ/+), and 6.5 hr (jbp2Δ/Δ). Note that, in contrast to the SET27 knockout, deletion of JBP2 was made via CRISPR/Cas9 in a cell line with integrated pJ1339 plasmid (see Materials and methods). (C) Tracks showing the distribution of RNA-seq reads in the presence (wild-type and jbp2Δ/+) or absence of JBP2 (jbp2Δ/Δ) around the same sTSR shown in Figure 4A (left). CRD1 ChIP (top track) is included to mark the position of SPARC sites. ORFs are indicated by grey boxes and directionality is shown with inset white arrows. Genes present within a single polycistron are connected with a thin black line.
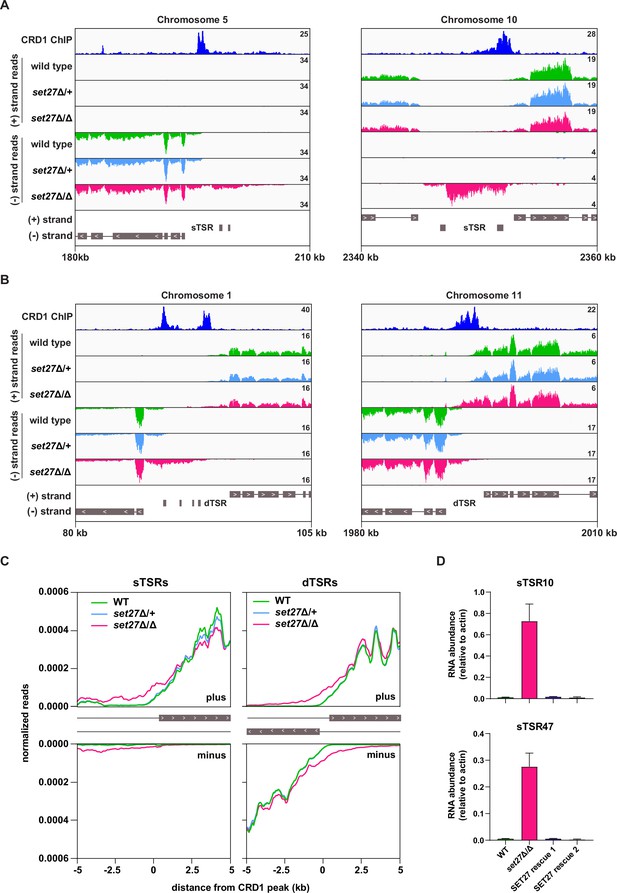
Transcription initiation is dysregulated in the absence of SET27.
(A) Tracks showing the distribution of RNA-seq reads in the presence (wild-type and set27Δ/+) or absence of SET27 (set27Δ/Δ) around selected unidirectional sTSR promoters. CRD1 ChIP (top track) is included to mark the position of SPARC sites. ORFs are indicated by gray boxes, and directionality is shown with inset white arrows. Genes present within a single polycistron are connected with a thin black line. Hypothetical protein-coding genes annotated within each TSR region are not connected to neighboring polycistrons. (B) As in (A), but for dTSR promoters. (C) Left: metaplots showing the distribution of RNA-seq reads around 33 SPARC sites marking unidirectional sTSR promoters. For clarity, we excluded SPARC sites present within 5 kb of a different SPARC site. Transcription in the forward direction is shown in the upper panel and transcription in the reverse direction is shown below. Right: metaplots showing the distribution of RNA-seq reads surrounding all 120 SPARC sites present at dTSR promoters. (D) RT-qPCR of RNA originating from sTSR10 and sTSR47 in wild-type, set27Δ/Δ, and two SET27 rescue clones. Error bars: standard deviation (SD) of two biological replicates.
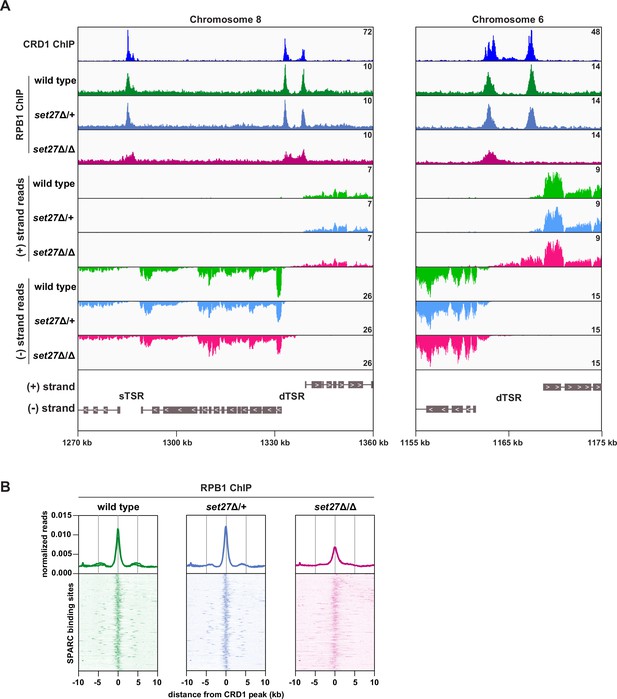
SET27 is required for full RNAPII recruitment to transcription start regions.
(A) Tracks showing the distribution of YFP-tagged RPB1, the largest subunit of T. brucei RNAPII, across selected genomic windows following ChIP-seq. CRD1 ChIP (top) and RNA-seq (bottom) tracks are included for comparison. (B) RPB1 enrichment profiles. CRD1 is used as a reference because it has the most prominent peaks at TSRs. The metagene plots (top) show normalized RPB1 read density around all SPARC sites, with each replicate plotted separately. The heatmaps (bottom) show RPB1 density around 177 individual SPARC sites. Each heatmap shows the average of at least two replicates.
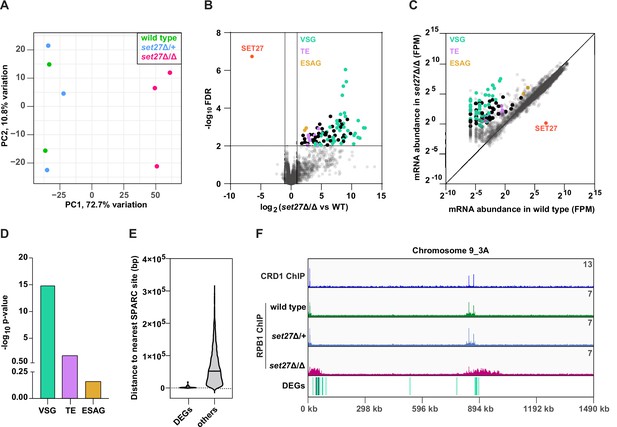
SET27 deletion derepresses VSGs and transposon-associated genes.
(A) Principal component analysis (PCA) comparing mRNA expression (n=8540) in wild-type, set27Δ/+, and set27Δ/Δ cells. (B) Volcano plot showing differentially expressed genes (DEGs) between wild-type and set27Δ/Δ cells. Cut-offs used for significance: log2 (tagged/untagged)>1 or <−1 and FDR<0.01. VSGs are colored in green; SLACS and ingi transposable element (TE) genes are in lavender; ESAG genes are in yellow; other DEGs are in black; and non-DEGs are in gray. See Supplementary file 3 for a complete list of DEGs. (C) Scatter plot of mRNA abundance normalized as FPM (fragments per million) in wild-type versus set27Δ/Δ cells. The diagonal marks the position of genes with equal expression in the wild-type and set27Δ/Δ cell lines. The color scheme is the same as in (B). (D) Gene ontology enrichment among upregulated mRNAs. P-values were calculated using Fisher’s exact test. (E) Violin plot showing the distance between the 5′ end of each DEG and the nearest SPARC site. Non-DEGs (others) are included as a comparison. Only genes found in the main body of chromosomes were included in this analysis. Subtelomeric genes were excluded because they frequently recombine and their precise location is uncertain. (F) The Chromosome 9_3A subtelomeric contig showing ChIP-seq reads for CRD1 and RPB1. The locations of DEGs are shown below. The color scheme is the same as in (B).
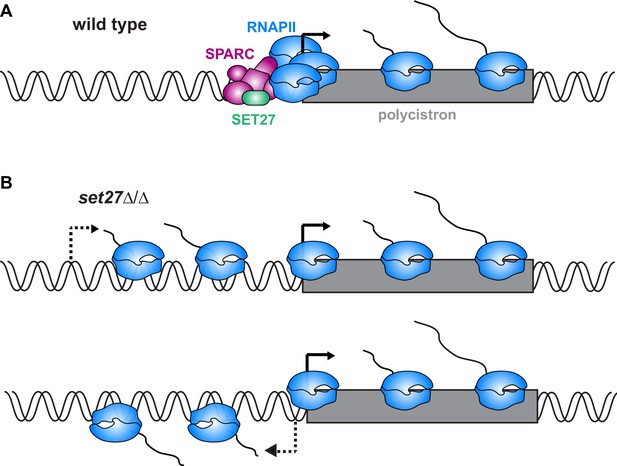
Model for SPARC-mediated definition of RNAPII transcription start sites.
(A) In wild-type cells, SPARC associates with genomic sites just upstream of polycistronic transcription units. SPARC sites coincide with promoters and regions of RNAPII enrichment. (B) In the absence of SET27, the SPARC complex dissociates from promoters and RNAPII enrichment is decreased. Transcription initiates upstream of the natural site (top) and sometimes in the reverse direction (bottom), effectively converting some unidirectional promoters into bidirectional promoters.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Trypanosoma brucei) | SET27 | TriTrypDB | Tb927.9.13470 | |
| Gene (T. brucei) | CRD1 | TriTrypDB | Tb927.7.4540 | |
| Gene (T. brucei) | CSD1 | This paper | Tb927.11.11840 | |
| Gene (T. brucei) | PHD6 | This paper | Tb927.3.2350 | |
| Gene (T. brucei) | PBP1 | This paper | Tb927.1.4250 | |
| Gene (T. brucei) | PWWP1 | This paper | Tb927.11.13820 | |
| Gene (T. brucei) | JBP2 | TriTrypDB | Tb927.7.4650 | |
| Gene (T. brucei) | RPB1 | TriTrypDB | Tb927.4.5020 | |
| Strain, strain background (T. brucei) | Lister 427 | Keith Matthews lab stocks | BF and PF | |
| Cell line (T. brucei) | YFP-SET27 | Staneva et al., 2021 | BF | |
| Cell line (T. brucei) | YFP-CRD1 | Staneva et al., 2021 | BF | |
| Cell line (T. brucei) | YFP-CSD1 | This paper | BF | |
| Cell line (T. brucei) | YFP-PHD6 | This paper | BF | |
| Cell line (T. brucei) | YFP-PBP1 | This paper | BF | |
| Cell line (T. brucei) | YFP-PWWP1 | This paper | BF | |
| Cell line (T. brucei) | YFP-JBP2 | This paper | BF | |
| Cell line (T. brucei) | YFP-SET27 | This paper | PF | |
| Cell line (T. brucei) | YFP-CRD1 | This paper | PF | |
| Cell line (T. brucei) | YFP-JBP2 | This paper | PF | |
| Cell line (T. brucei) | YFP-RPB1 | Staneva et al., 2021 | BF | |
| Cell line (T. brucei) | set27Δ/+ in YFP-RPB1 | This paper | BF | |
| Cell line (T. brucei) | set27Δ/Δ in YFP-RPB1 | This paper | BF | |
| Cell line (T. brucei) | jbp2Δ/+ in J1339 WT | This paper | BF | |
| Cell line (T. brucei) | jbp2Δ/Δ in J1339 WT | This paper | BF | |
| Cell line (T. brucei) | FLAG-CRD1 in WT | This paper | BF | |
| Cell line (T. brucei) | FLAG-CRD1 in set27Δ/Δ | This paper | BF | |
| Transfected construct (T. brucei) | BSR-YFP contruct | This paper | Used to generate the YFP-tagged cell lines | |
| Transfected construct (T. brucei) | PUR-FLAG construct | This paper | Used to generate the FLAG-tagged cell lines | |
| Transfected construct (T. brucei) | SET27/JBP2 5′ UTR - HYG/G418 - SET27/JBP2 3′ UTR | This paper | Used to generate the set27 and jbp2 Δ/+ and Δ/Δ cell lines | |
| Transfected construct (T. brucei) | SET27 5′ UTR - PUR - SET27 CDS - SET27 3′ UTR | This paper | Used to generate the SET27 rescue cell line | |
| Antibody | Anti-GFP (Rabbit polyclonal) | Thermo Fisher Scientific | Cat # A-11122 | IF (1:500) ChIP (10 µg per sample) |
| Antibody | Anti-GFP (Mouse monoclonal) | Roche | Cat # 11814460001 | IP-MS (10 µg per sample) |
| Antibody | M2 anti-FLAG (Mouse monosclonal) | Sigma-Aldrich | Cat #F1804 | IP-MS (10 µg per sample) |
| Antibody | Alexa Fluor 568 anti-rabbit (Goat polyclonal) | Thermo Fisher Scientific | Cat #A-11036 | IF (1:1000) |
| Recombinant DNA reagent | pPOTv4 BSR YFP (plasmid) | Dean et al., 2015 | Used for the YFP tagging | |
| Recombinant DNA reagent | pPOTv4 PUR FLAG (plasmid) | This paper | Used for the FLAG tagging | |
| Recombinant DNA reagent | pPOTv7 G418 mNG (plasmid) | Gift from Sam Dean | Used to generate the set27 and jbp2 Δ/+ and Δ/Δ cell lines | |
| Recombinant DNA reagent | pPOTv7 HYG RFP (plasmid) | Gift from Sam Dean | Used to generate the set27 and jbp2 Δ/+ and Δ/Δ cell lines | |
| Recombinant DNA reagent | pJ1339 (plasmid) | Alves et al., 2020 | Used to generate the jbp2 Δ/+ and Δ/Δ cell lines | |
| Recombinant DNA reagent | pMA-RQ-SET27 addback (plasmid) | Invitrogen | Used to generate the SET27 rescue cell line | |
| Commercial assay or kit | Quick Blunting Kit | NEB | Cat #E1201L | |
| Commercial assay or kit | Klenow Fragment (3′→5′ exo-) | NEB | Cat #M0212S | |
| Commercial assay or kit | NEXTflex-96 DNA Barcodes (Illumina Compatible) | Bioo Scientific | Cat #514105 | |
| Commercial assay or kit | Luna Universal One-Step RT-qPCR Kit | NEB | Cat #E3005S | |
| Commercial assay or kit | NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | NEB | Cat #7760 | |
| Commercial assay or kit | NEBNext Poly(A) mRNA Magnetic Isolation Module | NEB | Cat #E7490 | |
| Chemical compound, drug | AMPure XP beads | Beckman Coulter | Cat #A63881 | |
| Chemical compound, drug | TRIzol reagent | Thermo Fisher Scientific | Cat # 15596026 | |
| Chemical compound, drug | RapiGest SF Surfactant | Waters | Cat #186001861 | |
| Chemical compound, drug | Trypsin Protease, MS Grade | Thermo Fisher Scientific | Cat #90057 | |
| Software, algorithm | Cytoscape | Shannon et al., 2003 | ||
| Software, algorithm | LeishGEdit | Beneke et al., 2017 | ||
| Software, algorithm | Perseus | Tyanova et al., 2016 | ||
| Software, algorithm | TriTrypDB | Aslett et al., 2010 |
Additional files
-
Supplementary file 1
Significantly enriched interactors detected by proteomic analysis of affinity selected YFP- and FLAG-tagged proteins.
Cut-offs used to determine significant interacting partners: log2(tagged/untagged)>2 and P<0.01 (Student’s t-test).
- https://cdn.elifesciences.org/articles/83135/elife-83135-supp1-v2.xlsx
-
Supplementary file 2
Shared and unique interactors of affinity selected YFP-tagged proteins in BF T. brucei.
- https://cdn.elifesciences.org/articles/83135/elife-83135-supp2-v2.xlsx
-
Supplementary file 3
Differentially expressed mRNAs in set27Δ/Δ vs WT.
Cut-offs used to determine significant hits: fold change (FC) >2 or <-2 and false discovery rate (FDR)<0.01.
- https://cdn.elifesciences.org/articles/83135/elife-83135-supp3-v2.xlsx
-
Supplementary file 4
Oligos used in this study.
Upper case sequences align to T. brucei DNA. Lower case sequences align to pPOT plasmid DNA except for the sgRNA oligos targeting the JBP2 gene (see the "Notes" column for these oligos). "F" denotes a forward primer and "R" denotes a reverse primer.
- https://cdn.elifesciences.org/articles/83135/elife-83135-supp4-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83135/elife-83135-mdarchecklist1-v2.docx