Translation of dipeptide repeat proteins in C9ORF72 ALS/FTD through unique and redundant AUG initiation codons
Figures
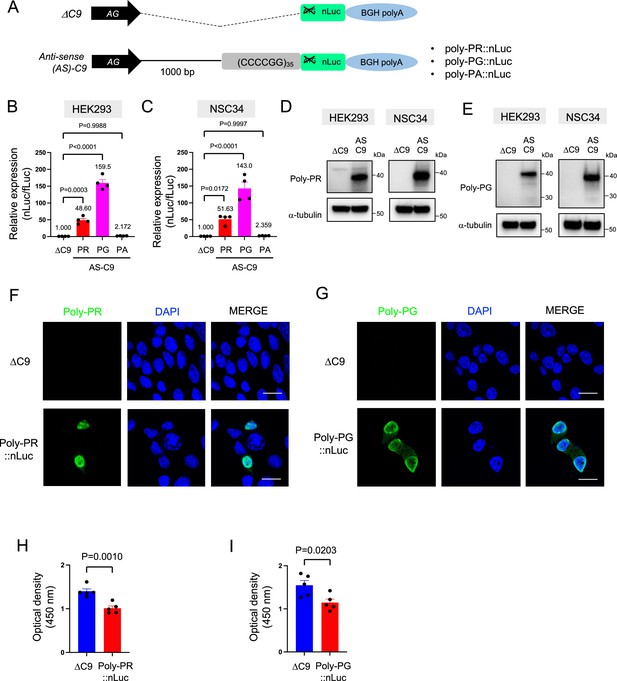
Poly-PR and poly-PG are translated from antisense CCCCGG repeats.
(A) Schematic diagram of the constructs with 35 CCCCGG repeats preceded by 1000-bp-long intronic sequence from human C9ORF72, and then followed by nanoluciferase (nLuc). (B) HEK293 and (C) NSC34 cells were cotransfected with fLuc along with either ΔC9 or AS-C9 plasmids. The levels of luciferase activity were assessed by dual luciferase assays (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed. (D–E) HEK293 and NSC34 cells were transfected with either ΔC9 or AS-C9 plasmids. Cell lysates were processed for western blotting, and immunostained with antibodies to (D) poly-PR, (E) poly-PG, and α-tubulin. (F–G) NSC34 cells transfected with either ΔC9, (F) poly-PR::nLuc, or (G) poly-PG::nLuc were stained with a nuclear marker (4′,6-diamidino-2-phenylindole [DAPI]: blue) and with antibodies against poly-PR (F: green) or poly-PG (G: green). Scale bars indicate 20 μm. (H–I) NSC34 cells were transfected with either ΔC9, (H) poly-PR::nLuc, or (I) poly-PG::nLuc plasmids. WST-8 assay was performed to assess the cell viability. The experiments were repeated five times. Unpaired t test was performed.
-
Figure 1—source data 1
Full raw unedited images of western blots shown in Figure 1.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig1-data1-v2.zip
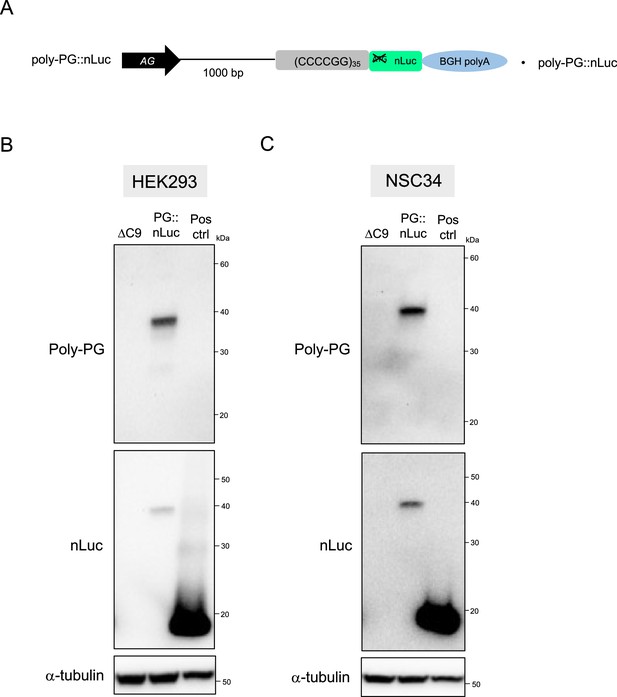
Nanoluciferase (nLuc) is fused to dipeptide repeats (DPRs) translated from antisense C9 plasmids containing CCCCGG repeats.
(A) Schematic diagram of the construct. (B) HEK293 and (C) NSC34 cells were transfected with either ΔC9, PG::nLuc, or positive control (Pos Ctrl) plasmids. The nanoluciferase (nLuc) expression plasmid pNL1.1.[Nluc/CMV] from Promega was used as positive control. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PG, nLuc, and α- tubulin.
-
Figure 1—figure supplement 1—source data 1
Full raw unedited images of western blots shown in Figure 1—figure supplement 1.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig1-figsupp1-data1-v2.zip
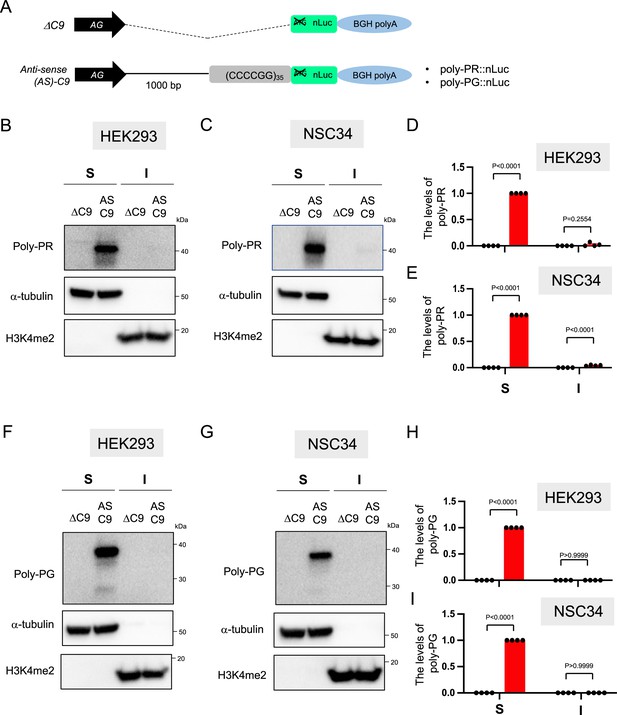
Expression levels of poly-PR and poly-PG in the RIPA-insoluble fraction.
(A) Schematic diagram of the constructs. (B, D, F, H) HEK293 and (C, E, G, I) NSC34 cells were transfected with either ΔC9 (blue) or AS-C9 (red) plasmids. Cell lysates were fractionated into RIPA-soluble (S) and -insoluble (I) fraction. Western blotting was performed and immunostained with antibodies to poly-PR (B–C), poly-PG (F–G), α-tubulin, and H3K4me2. H3K4me2 is a reliable marker for the RIPA-insoluble fraction (see Materials and methods). (D–E, H–I) The signal intensity of the bands was quantified. The expression levels of poly-PR (D–E) or poly-PG (H–I) in AS-C9 of RIPA-soluble fraction were set to 1.0. Experiments were repeated four times. Two-way ANOVA with Tukey’s multiple comparison test was performed.
-
Figure 1—figure supplement 2—source data 1
Full raw unedited images of western blots shown in Figure 1—figure supplement 2.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig1-figsupp2-data1-v2.zip
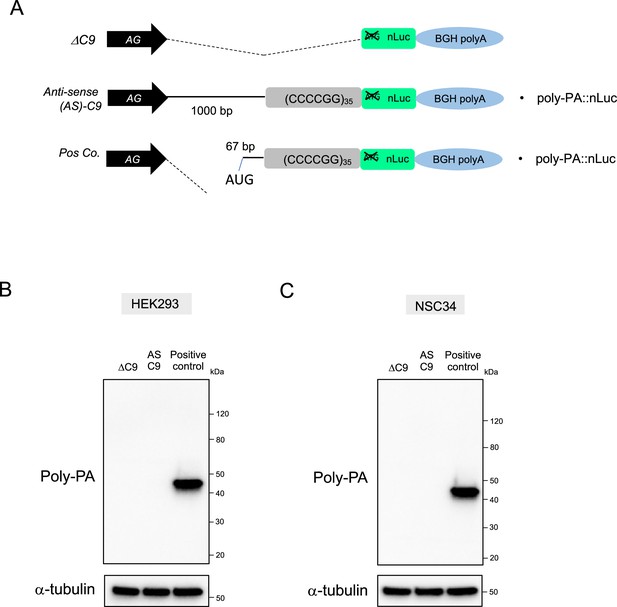
Poly-PA is not detected by western blotting upon transfection of antisense C9 plasmids containing CCCCGG repeats.
(A) Schematic diagram of the constructs. (B) HEK293 and (C) NSC34 cells were transfected with either ΔC9, AS-C9, or positive control plasmids. For positive control, we generated a plasmid that has an AUG start codon located at –67 bp upstream from CCCCGG repeats. This AUG is in poly-PA frame. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PA and α-tubulin.
-
Figure 1—figure supplement 3—source data 1
Full raw unedited images of western blots shown in Figure 1—figure supplement 3.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig1-figsupp3-data1-v2.zip
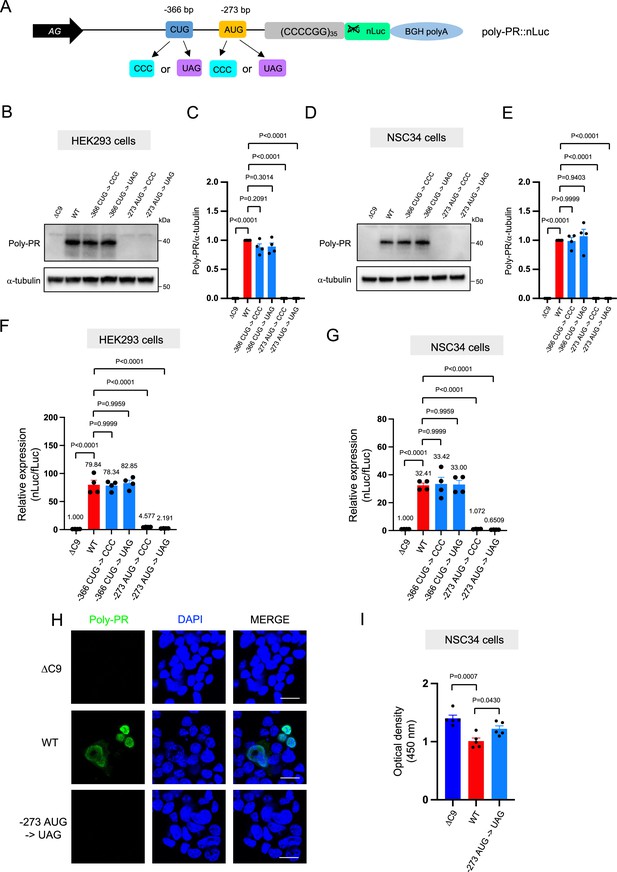
An AUG at –273 bp position is the start codon for poly-PR translation.
(A) Schematic diagram showing constructs with mutations in the putative start codons for poly-PR. HEK293 (B–C) and NSC34 (D–E) cells were transfected with indicated plasmids. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PR and α -tubulin. (B, D) Representative blots are shown. (C, E) The signal intensity of the bands were quantified (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed. (F) HEK293 and (G) NSC34 cells were cotransfected with the plasmids along with fLuc. The levels of luciferase activity were assessed by dual luciferase assays (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed. (H) NSC34 cells transfected with either ΔC9, poly-PR::nLuc, or –273 AUG ->UAG plasmids were stained with 4′,6-diamidino-2-phenylindole [DAPI] (blue) and immunostained with a poly-PR antibody (green). Scale bars show 20 μm. (I) NSC34 cells were transfected with either ΔC9, wild type (WT), or –273 AUG ->UAG plasmids. WST-8 assay was performed to assess the cell viability. The experiments were repeated five times. One-way ANOVA with Tukey’s multiple comparison test was performed. In ΔC9 and WT, the same datasets as Figure 1H were used (mean ± s.e.m.). The experiments were repeated five times. One-way ANOVA with Tukey’s multiple comparison test was performed.
-
Figure 2—source data 1
Full raw unedited images of western blots shown in Figure 2.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig2-data1-v2.zip
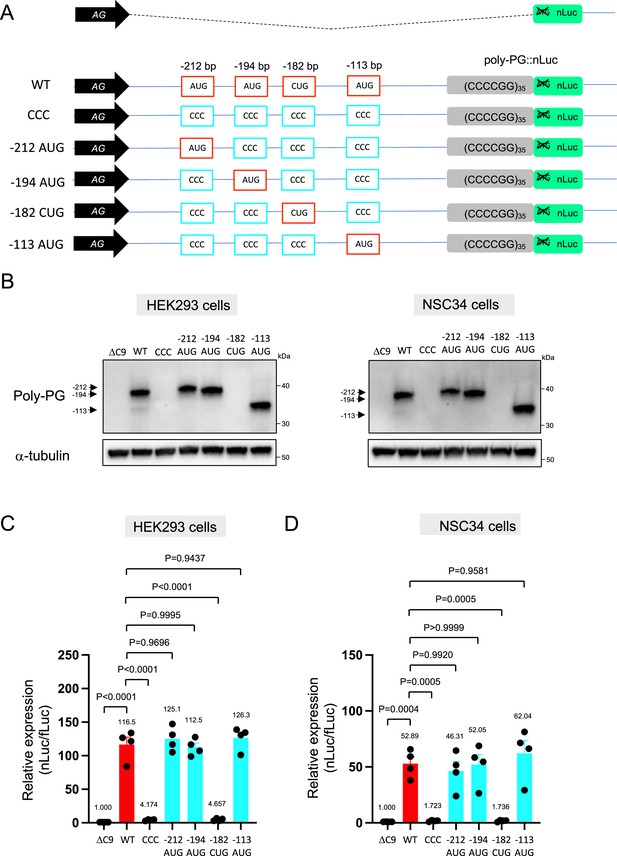
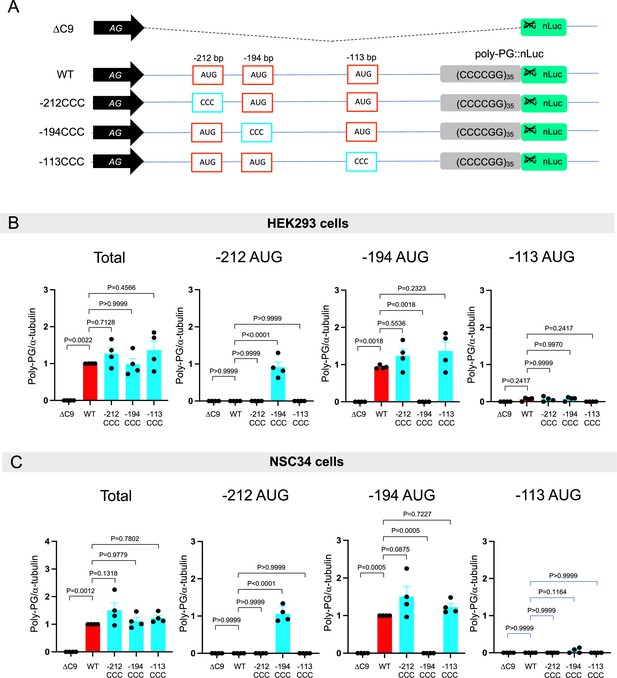
Mutation of AUG codons to CCC fails to suppress poly-PG translation.
(A) Schematic diagram showing mutants with changes in the putative start codons for poly-PG. (B) HEK293 and NSC34 cells were transfected with indicated plasmids. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PG and α-tubulin. (C) HEK293 and (D) NSC34 cells were cotransfected with fLuc plasmid along with other indicated plasmids. The level of luciferase activity was assessed by dual luciferase assay (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed.
-
Figure 3—source data 1
Full raw unedited images of western blots shown in Figure 3.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig3-data1-v2.zip
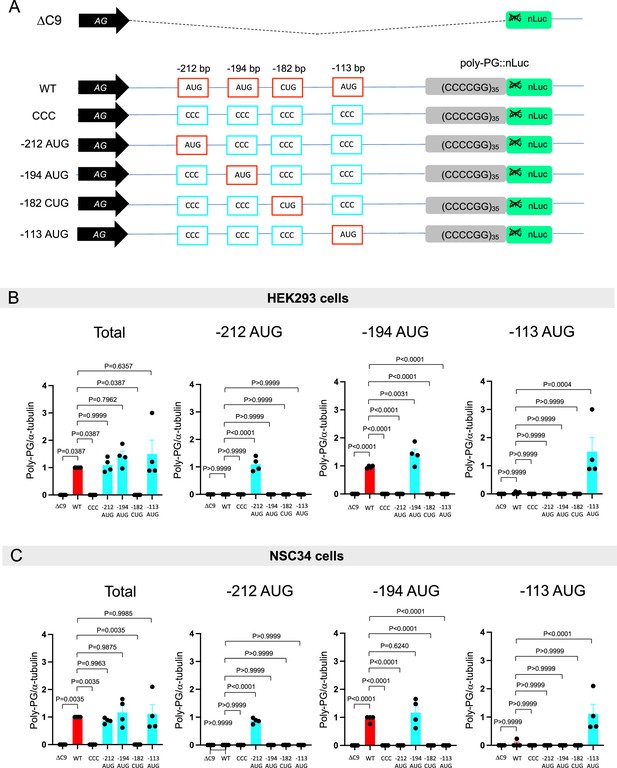
Quantification of data from Figure 3B.
(A) Schematic diagram of the constructs. (B–C) The signal intensity of total poly-PG and poly-PG translated from each of the AUGs (−212,–194, or –113) in (B) HEK293 and (C) NSC34 was quantified. One-way ANOVA with Tukey’s multiple comparison test was performed. The experiments were repeated four times. Data are presented as mean ± s.e.m.
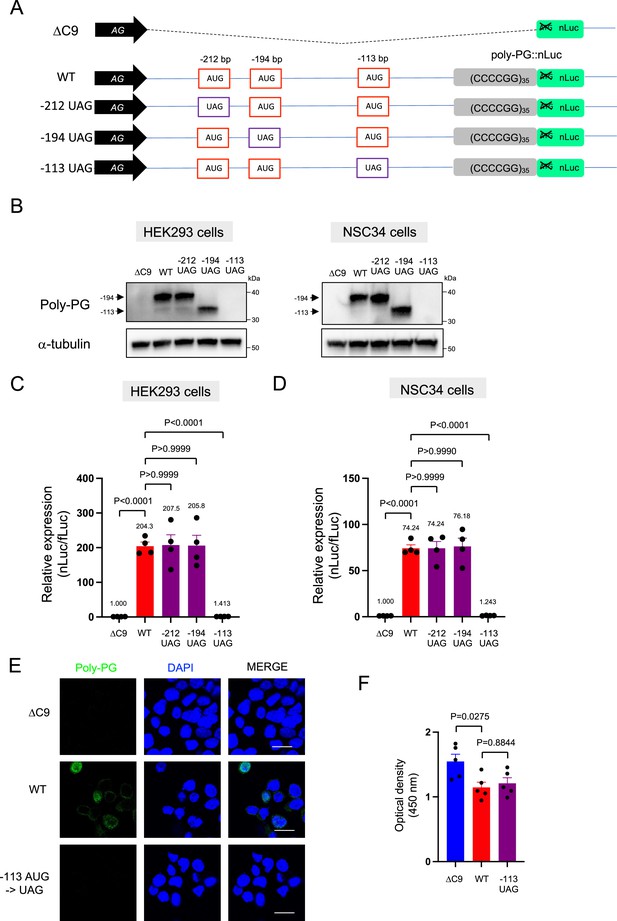
An AUG at –194 bp position is the primary start codon for poly-PG translation.
(A) Schematic diagram of the constructs. (B) HEK293 and NSC34 cells were transfected with indicated plasmids. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PG and α-tubulin. (C) HEK293 and (D) NSC34 cells were cotransfected with fLuc plasmid along with indicated plasmids. The level of luciferase activity was assessed by dual luciferase assays (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed.
-
Figure 4—source data 1
Full raw unedited images of western blots shown in Figure 4.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig4-data1-v2.zip
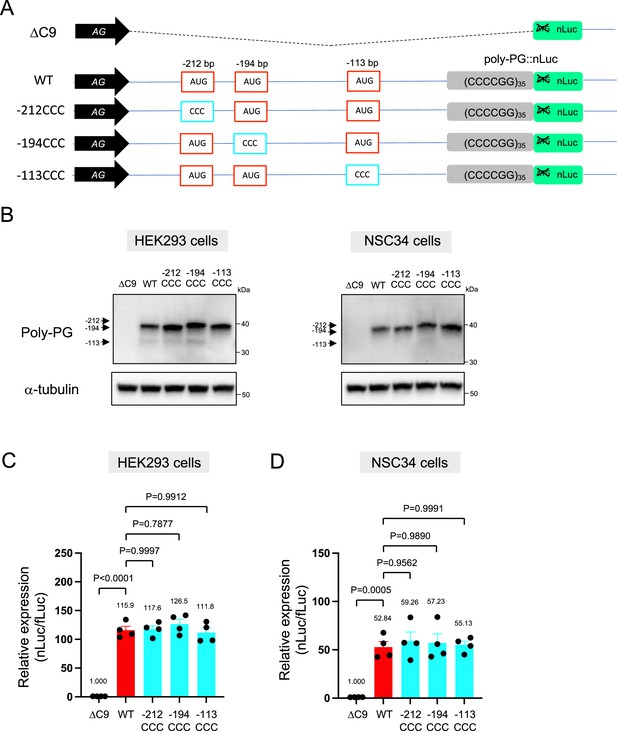
Quantification of data from Figure 4B.
(A) Schematic diagram of the constructs. (B–C) The signal intensity of poly-PG and poly-PG translated from each of the AUGs (−212, –194, or –113) in (B) HEK293 and (C) NSC34 cells was quantified. One-way ANOVA with Tukey’s multiple comparison test was performed. The experiments were repeated four times. Data are presented as mean ± s.e.m.
Redundancy of start codon usage in poly-PG translation.
(A) Schematic diagram of the constructs. (B) HEK293 and NSC34 cells were transfected with indicated plasmids. Cell lysates were processed for western blotting, and immunostained with antibodies to poly-PG and α-tubulin. (C) HEK293 and (D) NSC34 cells were cotransfected with fLuc plasmid along with indicated plasmids. The level of luciferase activity was assessed by dual luciferase assays (mean ± s.e.m.). The experiments were repeated four times. One-way ANOVA with Tukey’s multiple comparison test was performed. (E) NSC34 cells transfected with indicated plasmids were stained with 4′,6-diamidino-2-phenylindole [DAPI] (blue) and immunostained with a poly-PG antibody (green). Scale bars show 20 μm. (F) NSC34 cells were transfected with indicated plasmids. WST-8 assay was performed to assess the cell viability (mean ± s.e.m.). The experiments were repeated five times. One-way ANOVA with Tukey’s multiple comparison test was performed. In ΔC9 and wild type (WT), the same datasets as Figure 1I were used.
-
Figure 5—source data 1
Full raw unedited images of western blots shown in Figure 5.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig5-data1-v2.zip
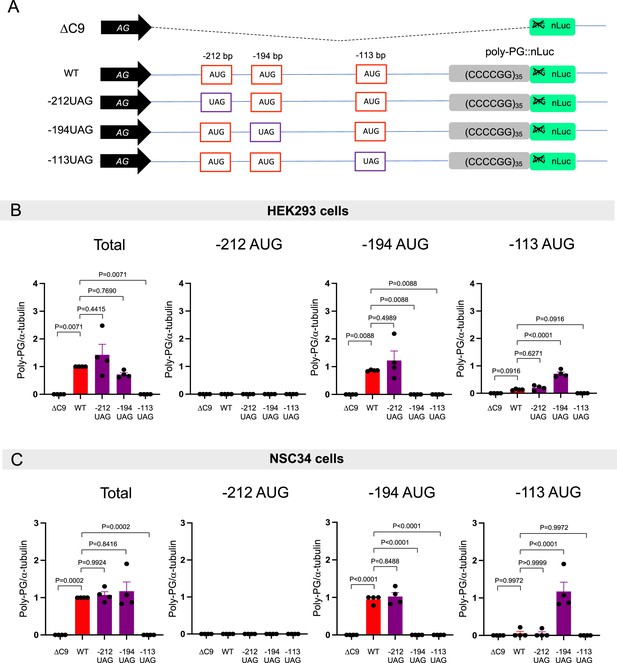
Quantification of data from Figure 5B and C.
(A) Schematic diagram of the constructs. (B–C) The signal intensity of total poly-PG and poly-PG translated from each of the AUGs (−212, –194, or –113) in (B) HEK293 and (C) NSC34 cells was quantified. One-way ANOVA with Tukey’s multiple comparison test was performed. The experiments were repeated four times. Data are presented as mean ± s.e.m.
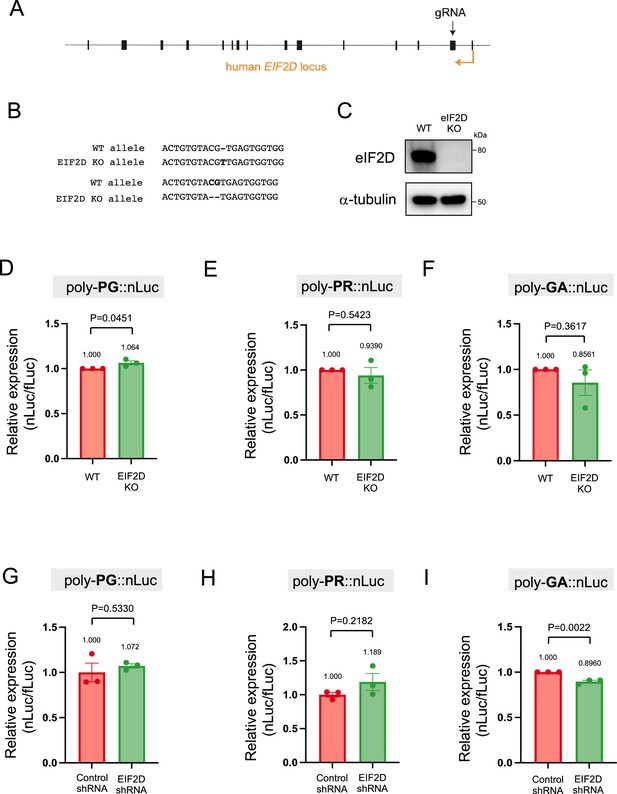
Downregulation of EIF2D does not reduce expression levels of poly-PG and poly-PR.
(A) A gRNA targeted the second exon of human EIF2D (see Materials and methods). (B) After CRISPR/Cas9-mediated gene editing, the EIF2D knockout (EIF2DKO) HEK293 cells carried different mutations on each allele. (C) Cell lysates from wild type (WT) and EIF2DKO HEK293 cells were processed for western blotting, and immunostained with antibodies to eIF2D and α-tubulin. (D–F) WT and EIF2DKO HEK293 cells were cotransfected with fLuc plasmid along with either (D–E) AS-C9 plasmids or (F) C9 plasmids containing 75 GGGGCC repeats. The level of luciferase activity was assessed by dual luciferase assays. (G–I) WT HEK293 cells were transfected with fLuc and either (G–H) AS-C9 plasmids or (I) C9 monocistronic plasmids containing 75 GGGGCC repeats along with anti-EIF2D short hairpin RNA (shRNA). The level of luciferase activity was assessed by dual luciferase assays (mean ± s.e.m.). The experiments were repeated three times. Unpaired t test was performed. The poly-GA reduction upon EIF2D shRNA is consistent with our previous observations (Sonobe et al., 2021), albeit more modest - likely due to a technical reason (a bicistronic construct containing 75 GGGGCC repeats was used in Sonobe et al., 2021).
-
Figure 6—source data 1
Full raw unedited images of western blots shown in Figure 6.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig6-data1-v2.zip
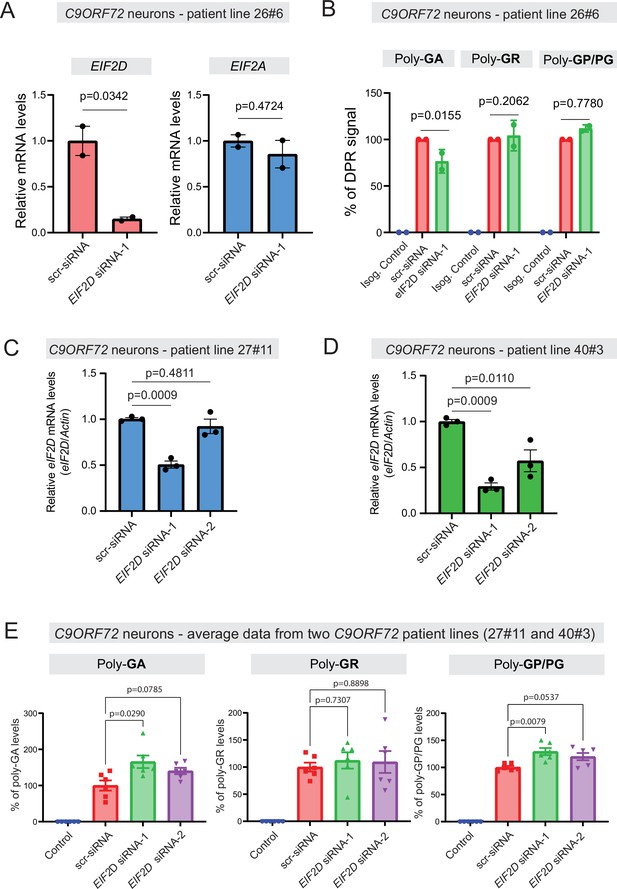
Dipeptide repeat (DPR) levels in human iPSC-derived neurons upon eIF2D knockdown.
(A) The EIF2D, EIF2A, and actin mRNA levels were assessed by real-time quantitative PCR on either isogenic control (26Z90) or C9ORF72 human motor neurons (patient line 26#6) upon small interfering RNA (siRNA) transfection (scramble or EIF2D siRNA-1). The eIF2D and eIF2A mRNA levels were normalized to actin. The experiments were repeated twice. p<0.05 by one-way ANOVA with Tukey’s post hoc test. (B) Poly-GA, poly-GR, and poly-GP levels in motor neurons differentiated independently (twice) from isogenic control and one C9ORF72 iPSC line. DPR levels were measured using an Meso Scale Discovery (MSD) immunoassay in a blinded manner. Data presented as mean ± SD. p-Values were calculated using two-way ANOVA with Dunnett’s multiple comparison test using Prism (9.1) software. (C–D) The EIF2D and actin mRNA levels were assessed by real-time quantitative PCR on C9ORF72 human motor neurons (two patient lines) upon siRNAs transfection (scramble, EIF2D siRNA-1 or EIF2D siRNA-2). The eIF2D mRNA levels were normalized to actin. The experiments were repeated three times. *p<0.05, ***p<0.001, ns, not significant by two-tailed unpaired t tests were used for two groups and a one-way ANOVA followed by Dunnett’s post hoc analysis was used for more than two groups. (E) Poly-GA, poly-GR, and poly-GP levels in motor neurons differentiated independently (n=3 times) from isogenic or healthy control lines and total two C9ORF72 patient iPSC lines (lines 27#11 and 40#3). DPR levels were measured using an MSD immunoassay in a blinded manner. For poly(GA) assay, total protein normalized poly(GA) concentrations were converted to percentage and presented as mean ± SE. For poly(GR), poly(GP) assay, total protein normalized electrochemiluminescence (ECL) values were converted to percentage and presented as mean ± SE. p-Values were calculated using one-way ANOVA with Dunnnett’s T3 multiple comparisons test .
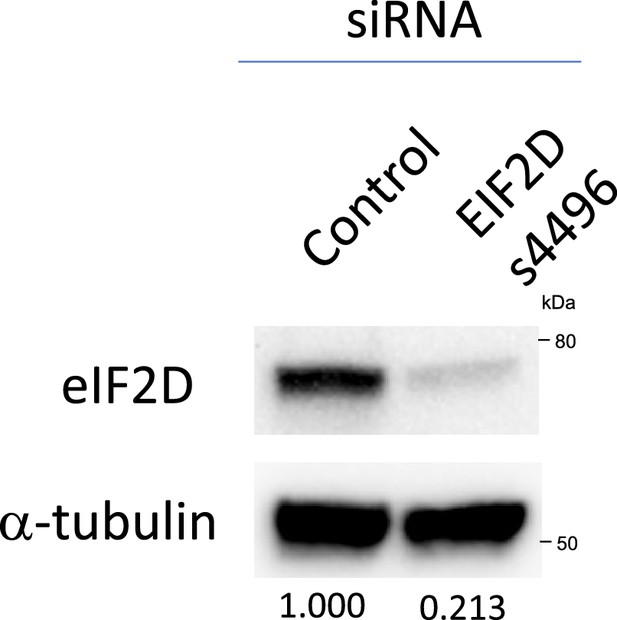
The small interfering RNA (siRNA) against eIF2D knocks down eIF2D protein levels.
siRNA (s4496) against eIF2D knocks down eIF2D protein levels. 3×105 HEK293 cells were plated into six-well cell culture plate. 60 pmol siRNAs were transfected into the cells using 6.25 μl Lipofectamine RNAiMAX. 72 hr later, protein was extracted using RIPA buffer and western blotting was performed.
-
Figure 7—figure supplement 1—source data 1
Full raw unedited images of western blots shown in Figure 7—figure supplement 1.
Figures with the uncropped blots are clearly labeled with the relevant bands.
- https://cdn.elifesciences.org/articles/83189/elife-83189-fig7-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 | ATCC | CRL-1573 | |
| Cell line (Mus musculus) | NSC34 | Gift from Dr. Neil R. Cashman (McGill University) PMID:1467557 | ||
| Cell line (Homo sapiens) | Isogenic iPS cells | Lopez-Gonzalez et al., 2019 PMID:31019093 | 26z90 | Isogenic control for patient line C926#6 |
| Cell line (Homo sapiens) | Isogenic iPS cells | Lopez-Gonzalez et al., 2019 PMID:31019093 | 27m91 | Isogenic control for patient line C927#11 |
| Cell line (Homo sapiens) | Healthy control iPS cells | Almeida et al., 2013 PMID:23836290 | Control2#20 | Control for patient line C940#3 |
| Cell line (Homo sapiens) | C9orf72 patient iPS cells | Almeida et al., 2013 PMID:23836290 | C926#6 | C9orf72 patient line |
| Cell line (Homo sapiens) | C9orf72 patient iPS cells | Almeida et al., 2013 PMID:23836290 | C927#11 | C9orf72 patient line |
| Cell line (Homo sapiens) | C9orf72 patient iPS cells | Freibaum et al., 2015 PMID:26308899 | C940#3 | C9orf72 patient line |
| Antibody | Anti-Poly-PR (Rabbit polyclonal) | EMD Millipore | ABN1354 | WB (1:1000) IF (1:250) |
| Antibody | Anti-Poly-PG (Mouse monoclonal) | Target ALS | TALS828.179 | WB (1:1000) IF (1:100) |
| Antibody | Anti-Poly-PA (Rabbit polyclonal) | EMD Millipore | ABN1356 | WB (1:1000) |
| Antibody | Anti-nLuc (Mouse monoclonal) | Promega | N700A | WB (1:500) |
| Antibody | Anti-α-tubulin (Rat monoclonal) | Abcam | Ab6160 | WB (1:5000) |
| Antibody | Anti-H3K4me2 (Rabbit polyclonal) | EMD Millipore | 07-030 | WB (1:2000) |
| Antibody | Anti-mouse horseradish peroxidase-conjugated secondary antibody (Sheep monoclonal) | GE Healthcare | NA931V | WB (1:5000) |
| Antibody | Anti-rabbit horseradish peroxidase-conjugated secondary antibody (Donkey monoclonal) | GE Healthcare | NA934V | WB (1:5000) |
| Antibody | Anti-rat horseradish peroxidase-conjugated secondary antibody (Goat polyclonal) | Cell Signaling Technology | 7077S | WB (1:1000) |
| Antibody | Alexa 488-conjugated anti-mouse IgG (Chicken polyclonal) | Thermo Fisher Scientific | A-21200 | IF (1:2000) |
| Antibody | Alexa 488-conjugated anti-rabbit IgG (Goat polyclonal) | Thermo Fisher Scientific | A-11008 | IF (1:2000) |
| Recombinant DNA reagent | pAG-ΔC9::nLuc | PMID:29792928 | ||
| Recombinant DNA reagent | pAG-AS(C9)-Poly-PR::nLuc (Plasmid) | This paper | Plasmid vector containing 35 CCCCGG repeats preceded by 1000-bp-long intronic sequence from human C9ORF72, and NanoLuc in frame of poly-PR | |
| Recombinant DNA reagent | pAG-AS(C9)-Poly-PG::nLuc (Plasmid) | This paper | Plasmid vector containing 35 CCCCGG repeats preceded by 1000-bp-long intronic sequence from human C9ORF72, and NanoLuc in frame of poly-PG | |
| Recombinant DNA reagent | pAG-AS(C9)-Poly-PA::nLuc (Plasmid) | This paper | Plasmid vector containing 35 CCCCGG repeats preceded by 1000-bp-long intronic sequence from human C9ORF72, and NanoLuc in frame of poly-PA | |
| Recombinant DNA reagent | pAG-AS(C9) -366CUG->CCC-Poly-PR::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PR::nLuc vector with mutation of the CTG at –366 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -366CUG->UAG-Poly-PR::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PR::nLuc vector with mutation of the CTG at –366 bp from CCCCGG repeats to TAG | |
| Recombinant DNA reagent | pAG-AS(C9) -273AUG->CCC-Poly-PR::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PR::nLuc vector with mutation of the ATG at –273 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -273AUG->UAG-Poly-PR::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PR::nLuc vector with mutation of the ATG at –273 bp from CCCCGG repeats to TAG | |
| Recombinant DNA reagent | pAG-AS(C9)CCC-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp, ATG at –194 bp, CTG at -182 bp, and ATG at –113 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -212AUG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –194 bp, CTG at –182 bp, and ATG at –113 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -194AUG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp, CTG at –182 bp, and ATG at –113 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -182CUG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp, ATG at –194 bp, and ATG at –113 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -113AUG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp, ATG at –194 bp, and CTG at –182 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -212CCC-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -194CCC-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –194 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -113CCC-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –113 bp from CCCCGG repeats to CCC | |
| Recombinant DNA reagent | pAG-AS(C9) -212UAG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –212 bp from CCCCGG repeats to TAG | |
| Recombinant DNA reagent | pAG-AS(C9) -194UAG-Poly-PG::nLuc | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –194 bp from CCCCGG repeats to TAG | |
| Recombinant DNA reagent | pAG-AS(C9) -113UAG-Poly-PG::nLuc (Plasmid) | This paper | pAG-AS(C9)-Poly-PG::nLuc vector with mutation of ATG at –113 bp from CCCCGG repeats to TAG | |
| Recombinant DNA reagent | lentiCRISPR v2-EIF2D (Plasmid) | This paper | Addgene (#52961) | lentiCRISPR plasmid containing gRNA sequence against EIF2D |
| Recombinant DNA reagent | Sh-Control (Plasmid) | PMID:34654821 | Thermo Fisher Scientific (#AM5764) | pSilencer 2.1-U6 neo plasmid containing non-specific control shRNA sequence |
| Recombinant DNA reagent | Sh-EIF2D (Plasmid) | PMID:34654821 | Thermo Fisher Scientific (#AM5764) | pSilencer 2.1-U6 neo plasmid containing shRNA sequence against EIF2D |
| Recombinant DNA reagent | pGL4.50 [luc2/CMV/ Hygro] (Plasmid) | Promega | E131A | Expression of firefly luciferase |
| Recombinant DNA reagent | pNL1.1 CMV (Plasmid) | Promega | N109A | Expression of NanoLuc |
| Recombinant DNA reagent | pcDNA 6/V5-His A (Plasmid) | Thermo Fisher Scientific | 43-0003 | |
| Sequence-based reagent | siRNA: non-targeting negative control | Thermo Fisher Scientific | 4390844 | Silencer Select |
| Sequence-based reagent | siRNA: EIF2D | Thermo Fisher Scientific | S4495 | Silencer Select |
| Sequence-based reagent | siRNA: EIF2D | Thermo Fisher Scientific | S4496 | Silencer Select |
| Chemical compound, drug | Halt Protease Inhibitor Cocktail | Thermo Fisher Scientific | 87786 | |
| Chemical compound, drug | SB421542 | Stemgent | 04-0010-10 | Neuron differentiation |
| Chemical compound, drug | CHIR99021 | Stem Cell Technologies | 72054 | Neuron differentiation |
| Chemical compound, drug | DMH1 | Stem Cell Technologies | 73634 | Neuron differentiation |
| Chemical compound, drug | All-Trans Retinoic Acid | Stem Cell Technologies | 72262 | Neuron differentiation |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0554S | |
| Commercial assay or kit | Nano-Glo Dual-Luciferase Reporter assay system | Promega | N1610 | |
| Commercial assay or kit | Cell Counting Kit-8 | Dojindo | CK-04 | |
| Commercial assay or kit | BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 | |
| Commercial assay or kit | 660 nm Protein Assay Reagent | Thermo Fisher Scientific | 22660 | |
| Software, algorithm | Image Lab software | Bio-Rad | ||
| Software, algorithm | ImageJ2 software | PMID:22930834 | ||
| Software, algorithm | GraphPad Prism | Dotmatics | ||
| Other | 5× passive lysis buffer | Promega | E1941 | Lysis buffer for luciferase assay |
| Other | 4′,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | D1306 | Nuclear staining (1 mg/ml) |
| Other | SuperSignal West Dura Extended Duration Substrate | Thermo Fisher Scientific | 34076 | Horseradish peroxidase substrate for western blotting |
| Other | Lipofectamine LTX | Thermo Fisher Scientific | 15338030 | Plasmid transfection reagent |
Additional files
-
Supplementary file 1
List of primers used for this study.
- https://cdn.elifesciences.org/articles/83189/elife-83189-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/83189/elife-83189-transrepform1-v2.pdf





