Augmin prevents merotelic attachments by promoting proper arrangement of bridging and kinetochore fibers
Figures
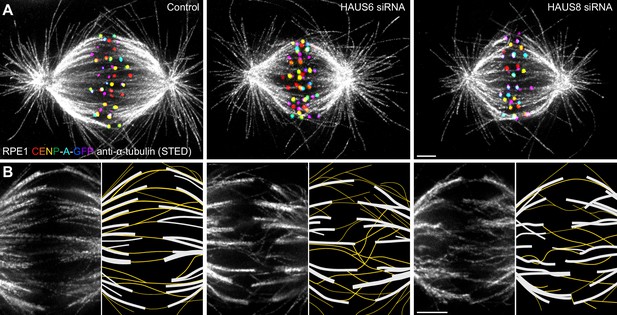
Augmin ensures the proper formation of entities consisting of bridging fibers that connect two sister k-fibers.
(A) STED superresolution images of microtubules immunostained for α-tubulin (gray) in control (left), HAUS6- (middle), and HAUS8-depleted (right) RPE1 cells stably expressing CENP-A-GFP (rainbow, confocal). Images show maximum intensity projections of 6 central z-planes of metaphase spindles. Kinetochores are color-coded for depth from blue to red with the Spectrum LUT in ImageJ. (B) Insets of STED superresolution images of microtubules (gray) in spindle midzones of control (left), HAUS6- (middle) and HAUS8-depleted (right) cells. Next to each image is the schematic representation of the microtubules in the midzone, with white lines representing k-fiber microtubules and yellow lines representing midplane-crossing microtubules. Images show a single z-plane and do not correspond to midzones of spindles in panel (A). All images are adjusted for clarity based on the intensity of astral microtubules in each image (see Materials and methods). Scale bars, 2 µm.
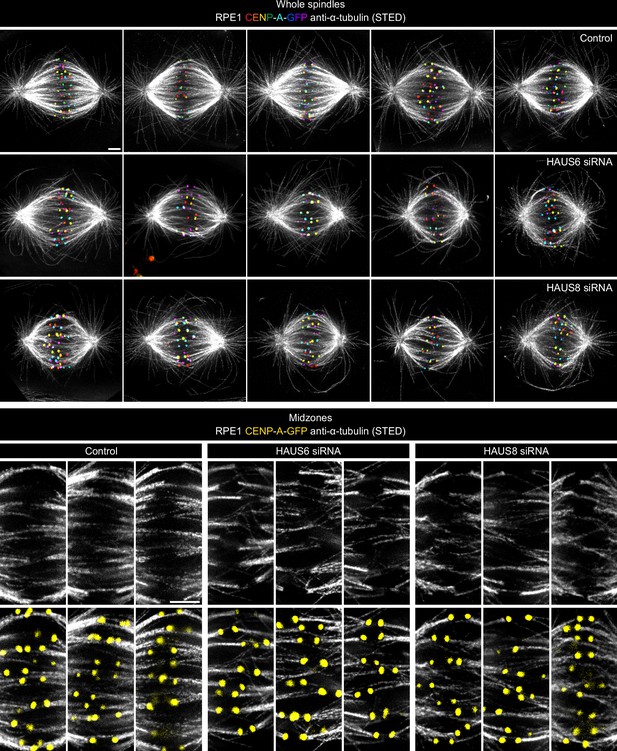
Augmin is necessary for the formation of uniformly arranged microtubule bundles in the spindle.
Panels with whole spindles: STED superresolution images of microtubules immunostained for α-tubulin (gray) in control (top), HAUS6- (middle), and HAUS8-depleted (bottom) RPE1 cells stably expressing CENP-A-GFP (rainbow, confocal). Images show maximum intensity projections of 6 central z-planes of metaphase spindles. The kinetochores are color-coded for depth from blue to red with the Spectrum LUT in ImageJ. Panels with midzones: insets of STED superresolution images of microtubules (gray) from spindle midzones in control (left), HAUS6- (middle), and HAUS8-depleted (right) RPE1 cells stably expressing CENP-A-GFP (yellow). Top row shows only microtubules present in the midzone area and bottom row shows the same midzones with the kinetochores. Images show single z-plane and do not correspond to midzones of spindles in the panel with whole spindles. All images are adjusted for clarity based on the intensity of astral microtubules in each image (see Materials and methods). Scale bars, 2 µm.
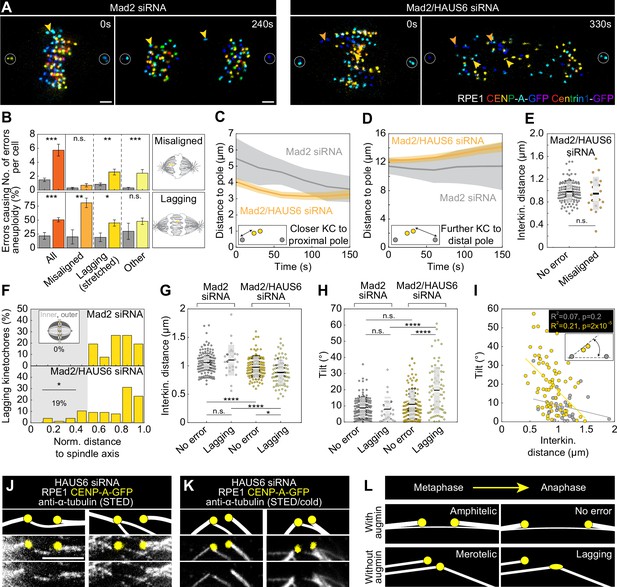
Augmin-nucleated midplane-crossing microtubules prevent kinetochore tilt and thus merotelic attachments.
(A) Time-lapse images of RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (rainbow, confocal) in Mad2-depleted cells (left) and Mad2/HAUS6-codepleted cells (right). Yellow arrows represent lagging kinetochores and orange arrows misaligned kinetochores. Kinetochores are color-coded for depth from blue to red with the 16 Colors LUT in ImageJ. (B) The number of segregation errors per cell (top) and the percentage of errors causing aneuploidy (bottom) in Mad2-depleted cells (gray) and Mad2/HAUS6-codepleted cells (dark orange, light orange, dark yellow, light yellow). All segregation errors (dark orange) are divided into three groups: misaligned (light orange), lagging (dark yellow) and other (light yellow). Schematic representations next to the graph represent misaligned kinetochores (top) and lagging kinetochore (bottom). The number of errors in Mad2-depleted cells - in total 46 errors in 22 out of 31 cells; 10 misaligned kinetochore pairs in 9 out of 31 cells; 26 lagging kinetochores in 16 out of 31 cells; 10 other errors in 9 out of 31 cells. Number of errors in Mad2/HAUS6-codepleted cells - in total 172 errors in 25 out of 30 cells; 21 misaligned kinetochore pairs in 11 out of 30 cells; 78 lagging kinetochores in 23 out of 30 cells; 73 other errors in 20 out of 30 cells. Aneuploidy in Mad2-depleted cells - in total 11/46 errors in 22 out of 31 cells; 2/10 misaligned kinetochore pairs in 9 out of 31 cells; 6/26 lagging kinetochores in 16 out of 31 cells; 3/10 other errors in 9 out of 31 cells. Aneuploidy in Mad2/HAUS6-codepleted cells - in total 47/172 errors in 25 out of 30 cells; 17/21 misaligned kinetochore pairs in 11 out of 30 cells; 35/78 lagging kinetochores in 23 out of 30 cells; 35/73 other errors in 20 out of 30 cells. (C) The distance of closer kinetochore to the proximal pole and (D) further kinetochore to the distal pole of misaligned kinetochore pairs in time for Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (orange). Values are shown as mean (dark line) and SEM (shaded areas). The insets show the positions of kinetochores (yellow) with respect to spindle poles (gray). (E) Univariate scatter plot of the interkinetochore distance of error-free kinetochore pairs (gray) and misaligned kinetochore pairs (orange) in Mad2/HAUS6-codepleted cells. N=30 cells and 120 error-free kinetochore pairs in Mad2-depleted cells and N=30 cells and 21 misaligned kinetochore pairs in Mad2/HAUS6-codepleted cells. (F) The percentage of lagging kinetochores in Mad2-depleted (top) and Mad2/HAUS6-codepleted cells (bottom) divided by their location with respect to the pole-to-pole axis into inner and outer (schematic representation shown as inset, see Materials and methods). (G) Univariate scatter plot of the interkinetochore distance of error-free and lagging kinetochore pairs in Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (yellow). (H) Univariate scatter plot of the angle that the error-free and lagging kinetochore pairs form with the pole-to-pole axis (tilt) in Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (yellow). N=31 cells and 124 error-free and 26 lagging kinetochore pairs from Mad2-depleted cells. N=30 cells and 120 error-free and 78 lagging kinetochore pairs from Mad2/HAUS6-codepleted cells. (I) The correlation of the tilt and the interkinetochore distance for Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (yellow). Inset shows schematic representation of the tilt (kinetochores are shown in yellow and spindle poles in gray). (J) The insets of kinetochore pairs with merotelic attachments in RPE1 cells stably expressing CENP-A-GFP (yellow, confocal) and immunostained for α-tubulin (gray, STED). (K) The insets of kinetochore pairs from cells as in (J) but exposed to cold treatment. Images in (J) and (K) are smoothed with 0.5-mm-sigma Gaussian blur and adjusted for clarity (see Materials and methods). Schematic representations in (J) and (K) are shown above the insets for better visualization of merotelic microtubule attachments. (L) The schematic representations of a kinetochore pair with amphitelic attachment in metaphase that does not cause any segregation errors during anaphase when augmin is present (top) and a kinetochore pair with merotelic attachment in metaphase that ends up as the lagging kinetochore during anaphase when augmin is not present (bottom). (E, G and H) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Statistical analysis (B (top) and E) Mann–Whitney U test; (B (bottom) and F) Fisher’s exact test; (G and H) ANOVA with the post-hoc Tukey test; (I) linear regression; p-value legend: <0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). Scale bars, 2 µm.
-
Figure 2—source data 1
Immunoblot analysis of HAUS6 siRNA treatment efficiency in RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP.
Full unedited blots of all three independent experiments are shown.
- https://cdn.elifesciences.org/articles/83287/elife-83287-fig2-data1-v2.pdf
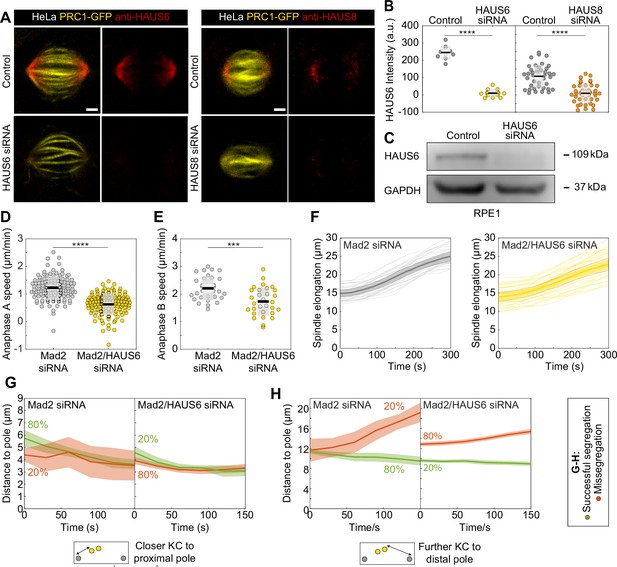
Augmin is required for accurate chromosome segregation during anaphase.
(A) Fixed control, HAUS6 and HAUS8 siRNA-treated HeLa cells stably expressing PRC1-GFP (yellow) and immunostained for HAUS6 (red, left panels) or HAUS8 (red, right panels). Chromosomes were stained with DAPI (not shown) to identify cells in metaphase. Images are sum intensity projections of nine central z-planes. (B) Univariate scatter plot of HAUS6 and HAUS8 intensities in control cells (gray) and cells depleted of HAUS6 (yellow) or HAUS8 (orange). The intensity of HAUS6 and HAUS8 in siRNA-treated cells is reduced by 96 ± 2% and 82 ± 16% compared to control cells, respectively. (C) Immunoblot analysis of HAUS6 siRNA treatment efficiency in RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP. HAUS6 antibody was used to validate the efficiency of knockdown, with GAPDH as the loading control. The representative image of three independent experiments is shown. (D) Univariate scatter plot of anaphase A speed, defined as the distance of kinetochore and the closer pole in time, for Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (yellow). The anaphase A speed in Mad2-depleted cells is 1.2±0.04 µm/min, and in Mad2/HAUS6 codepleted cells 0.6±0.03 µm/min. N=120 kinetochores from 30 cells in both treatments. (E) Univariate scatter plot of anaphase B speed, defined as the distance between two poles in time, for Mad2-depleted (gray) and Mad2/HAUS6-codepleted cells (yellow). The anaphase B speed in Mad2-depleted cells is 2.2±0.08 µm/min, and in Mad2/HAUS6-codepleted cells 1.7±0.09 µm/min. N=31 Mad2-depleted cells and N=30 Mad2/HAUS6-codepleted cells. (F) A plot of spindle elongation in time following Mad2 depletion (gray) and Mad2/HAUS6 (yellow) codepletion. (G) A plot of distance of the misaligned kinetochore that is closer to the proximal pole in time following Mad2 depletion (left) and Mad2/HAUS6 (right) codepletion. (H) A plot of distance of the misaligned kinetochore that is further away from the proximal pole in time following Mad2 depletion (left) and Mad2/HAUS6 (right) codepletion. Kinetochore pairs in (G) and (H) are divided based on their outcome in anaphase into those that successfully segregated (green) and those that missegregated (red). Insets show schematic representations of the distance of closer and further kinetochore (yellow) to their corresponding spindle pole (gray). (B, D and E) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). (F–H) Values are shown as mean (thick line) and SEM (shaded area). All results were obtained from three independent experiments. Statistical analysis (B and E) t-test; (D) Mann-Whitney U test; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). Scale bars, 2 µm.
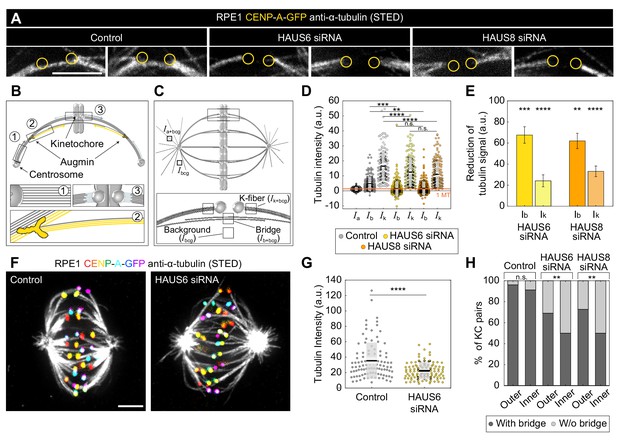
Augmin is crucial for the nucleation of bridging microtubules.
(A) The insets of kinetochore pairs in RPE1 cells stably expressing CENP-A-GFP (not shown) immunostained for α-tubulin (gray, STED) in control cells (left) and after HAUS6 (middle) or HAUS8 (right) depletion. The insets demonstrate kinetochore pairs with bridging fibers affected by HAUS6 or HAUS8 depletion compared to bridging fibers in control cells. The positions of kinetochores are marked with yellow circles. (B) The schematic representation of three possible pathways of microtubule nucleation: (1) centrosome-dependent (2) augmin-dependent and (3) chromatin- and kinetochore-dependent nucleation. The augmin complex is shown in yellow. (C) Top: the schematic representation of the mitotic spindle in metaphase and the method used to measure the tubulin intensity of the astral microtubules. Small square regions were measured on microtubules extending from the spindle pole, corresponding to astral microtubules. Their background was measured in the empty area between the two astral microtubules, and it was subtracted from astral microtubule intensity. Bottom: Schematic representation of the method used to measure the tubulin intensity of the bridging and k-fiber. Small square regions were measured between two kinetochores or right next to the kinetochore, corresponding to bridging and k-fibers, respectively. The intensity of k-fibers was measured as an average of two sister k-fibers, and the average value of the background within the spindle was subtracted from all measurements. Ia+bcg = intensity of astral microtubules with background, Ik+bcg = intensity of k-fibers with background, Ib+bcg = intensity of bridging microtubules with background, Ibcg = intensity of background. (D) Univariate scatter plot of tubulin signal intensities of astral microtubules in control cells (reference value, dark gray, Ia), and bridging fibers (Ib) and k-fibers (Ik) in control cells (gray), HAUS6- (yellow) and HAUS8-depleted cells (orange). (E) The reduction of tubulin signal in the bridging fiber (Ib) and the k-fiber (Ik) following HAUS6 (yellow) or HAUS8 (orange) depletion, values are shown as mean ± SEM. p-Values were calculated using the absolute values of tubulin signal intensity of bridging or k-fibers following HAUS6 or HAUS8 depletion, compared to the absolute values of tubulin signal intensity of corresponding fibers in control cells. (D) and (E) N=30 cells and 90 astral microtubules in control cells, 158 bridging and sister k-fibers in control and 180 bridging and sister k-fibers in HAUS6- and HAUS8 siRNA-treated cells. (F) STED superresolution images of microtubules stained for α-tubulin (gray) in RPE1 cells stably expressing CENP-A-GFP (rainbow, confocal) in control cells (left) and HAUS6 siRNA-treated cells (right) exposed to cold treatment. The images are maximum intensity projections and kinetochores are color-coded for depth from blue to red with the Spectrum LUT in ImageJ. (G) Univariate scatter plot of the tubulin signal intensities of k-fibers in control cells (gray) and upon HAUS6 depletion (yellow) in cells exposed to cold treatment. N=30 cells and 101 bundles in control cells and 102 bundles in HAUS6-depleted cells. (H) The fractions of kinetochore pairs with bridging fibers (dark gray) and with undetectable bridging fibers (light gray) in control, HAUS6- and HAUS8-depleted cells. Kinetochore pairs are divided based on their location in the spindle into outer and inner (See Materials and methods and Results). (D) and (G) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Statistical analysis (D and E) ANOVA with post-hoc Tukey test, (G) Mann–Whitney U test, (H) chi-square test; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). All images are adjusted for clarity (see Materials and methods). Scale bars, 2 µm.
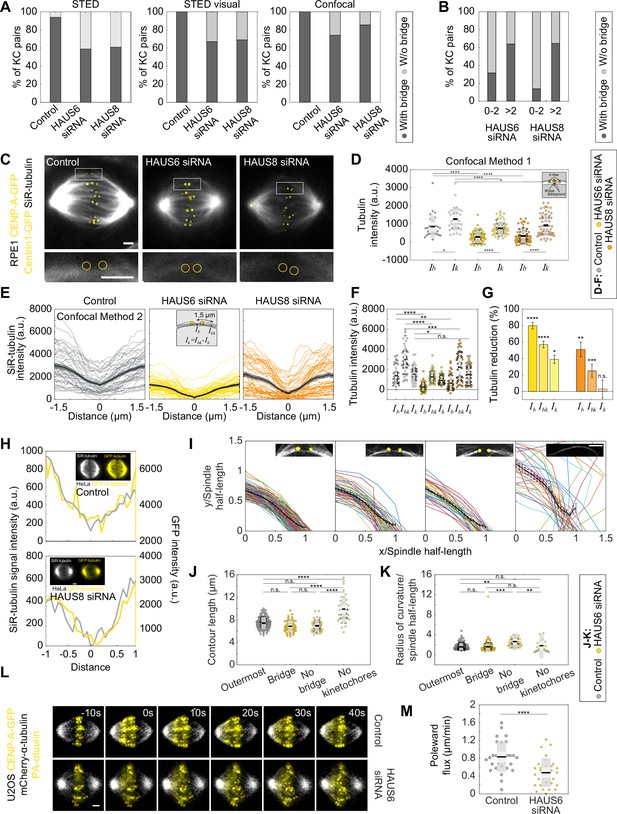
Augmin ensures proper architecture and dynamics of the metaphase spindle.
(A) The fractions of kinetochore pairs with bridging fibers (dark gray) and with undetectable bridging fibers (light gray). The fractions are obtained from the tubulin intensity measured in STED images (left), by visual inspection of STED images (middle) and from SiR-tubulin intensity measured in confocal images (right) of control, HAUS6- and HAUS8-depleted cells. (B) The fractions of kinetochore pairs with bridging fibers (dark gray) and without bridging fibers (light gray) depending on the projected distance from the long (pole-to-pole) spindle axis in confocally imaged HAUS6- (left) and HAUS8- (right) depleted cells. (C) Live images (single z-plane) of SiR-tubulin (gray) stained metaphase spindles in control (left), HAUS6- (middle) and HAUS8-depleted (right) RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (both in yellow). The enlarged boxes show bridging fibers affected by HAUS6 or HAUS8 depletion compared to the bridging fiber in control cell. The positions of kinetochores are marked with yellow circles. (D) Univariate scatter plot of the tubulin signal intensities of bridging and k-fibers in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP. Schematic representation of the method for measuring tubulin intensity of a bridging fiber and corresponding k-fibers (kinetochores are shown in yellow and tubulin signal in white). For the analysis, the average of two k-fibers is calculated and the average of background is subtracted from the tubulin intensity values of the bridging and k-fiber. N=30 bridging and sister k-fibers in 10 control cells and N=60 bridging and sister k-fibers in both 10 HAUS6- and 10 HAUS8-depleted cells. (E) Intensity profiles of SiR-tubulin signal along the bundles in control cells (gray) and after HAUS6 (yellow) or HAUS8 (orange) depletion. The center of the bridging fiber is set at a distance zero and the minimum intensity of the central part of the spindle was subtracted from the values of intensity profiles. N=10 cells and 50 bundles for control and HAUS6 siRNA-treated cells, N=10 cells and 48 bundles for HAUS8 siRNA-treated cells. Mean ± SEM (thick black line and shaded area, respectively). The method for measuring the tubulin intensity of the bridging fiber and the corresponding k-fibers is depicted in the inset; the average of two k-fibers was calculated whenever both k-fibers resided in the same-z plane. (F) Univariate scatter plot of the tubulin signal intensities of the bridging fiber (Ib), the bundle consisting of the k-fiber and bridging fiber (Ibk), and k-fiber (Ik) in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) cells. (G) Reduction of the tubulin signal in the bridging fiber (Ib), the bundle consisting of the k-fiber and bridging fiber (Ibk), and the k-fiber (Ik) following HAUS6 (yellow) or HAUS8 (orange) depletion. Values are shown as mean ± SEM. P-values were calculated using the absolute values of tubulin signal intensity of bridging or k-fibers following HAUS6 or HAUS8 depletion, compared to the absolute values of tubulin signal intensity of corresponding fibers in control cells. (H) Intensity profiles of SiR-tubulin (gray) and GFP-tubulin (yellow) signal in HeLa cells stably expressing GFP-α-tubulin (yellow) and stained with SiR-tubulin (gray). The values of two intensity profiles are adjusted to observe the level of overlapping between them and the minimum of the SiR-tubulin signal is set as a distance zero. This method was used as a control to validate SiR-tubulin labeling. (I) Bundle contours from RPE1 cells stably expressing CENP-A-GFP (yellow). Examples of each bundle type are shown in insets. From left to right: the outermost bundle in control cells, the outermost bundle with a bridging fiber, the outermost bundle without a bridging fiber and the outermost bundle without kinetochores in HAUS6-depleted cells; mean ± SEM is shown in black. (J) Univariate scatter plot of contour lengths and (K) radii of curvature in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) cells. N=120 outermost bundles from 30 control cells, 54 bundles with a bridging fiber, 40 bundles without a bridging fiber, and 36 bundles without kinetochores from 30 HAUS6-depleted cells. (L) Time-lapse images of control (top) and HAUS6-depleted (bottom) U2OS cells stably expressing CENP-A-GFP (yellow), mCherry-α-tubulin (gray) and PA-GFP-α-tubulin, before (–10 s), at the time when photoactivation (yellow) was performed (0 s), and after photoactivation. (M) Univariate scatter plot of the poleward flux speed in control cells (gray) and after HAUS6 depletion (yellow). N=30 measured photoactivation spots in 30 cells for both conditions. (D, F, J, K and M) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Images in (L) are adjusted for clarity (see Materials and methods). Statistical analysis; (D, F, G, J and K) ANOVA with post-hoc Tukey test; (M) t-test, p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). Scale bars, 2 µm.
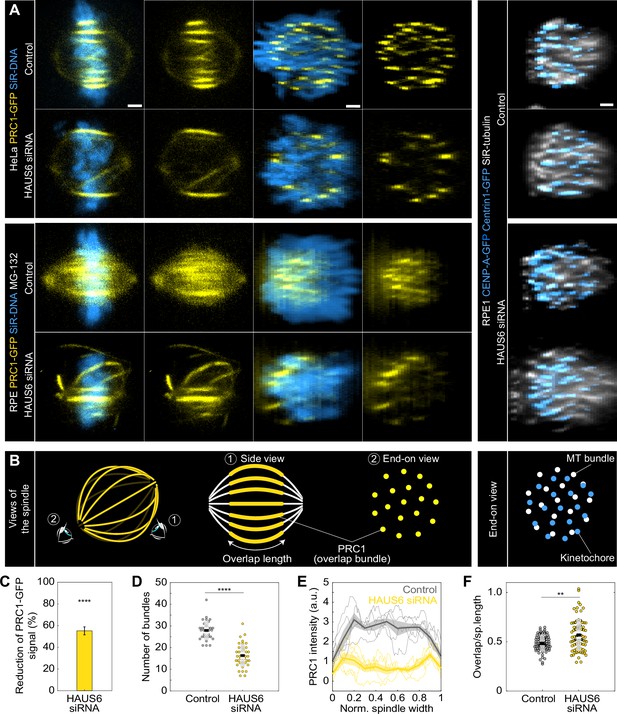
Augmin-depleted spindles have fewer bridging fibers, which have larger overlap length and are located at the spindle periphery.
(A) The four columns on the left represent live images of metaphase spindles in untreated HeLa or MG-132-treated RPE1 cells stably expressing PRC1-GFP (yellow) and stained with SiR-DNA (blue) in control cells (top rows) and after HAUS6 depletion (bottom rows). 1st and 2nd column: side view of the spindle; 3rd and 4th column: end-on view of the same spindle, showing a barrel-like arrangement of PRC1-labeled bundles after augmin depletion. Images on the right show the end-on view of RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (both in blue) and stained with SiR-tubulin (gray) in control cells (top) and after HAUS6 depletion (bottom). Side views are sum intensity projections of 5 central z-slices (∆z=0.5 µm) in HeLa cells and 10 central z-slices for RPE1 cells. End-on views are sum projections of 10 central z-slices (∆z=0.083 µm) for HeLa and 20 central z-slices for RPE1 cells. (B) Left: schematic representations of different views of the spindle. Eye signs mark the angle for the side view (1) and the end-on view (2). Side view was used to measure the length of overlap regions (yellow) and end-on view to determine the number of bundles (yellow dots). Right: schematic representation of the end-on view of RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (blue dots) and stained with SiR-tubulin (gray dots). (C) The reduction of the PRC1 signal in RPE1 cells treated with MG-132 measured in sum intensity projection of 10 central z-slices following HAUS6 depletion. Values are shown as mean ± SEM. P-values were calculated using the absolute values of PRC1 signal intensity following HAUS6 depletion (N=39 cells), compared to the absolute values of PRC1 signal intensity in control cells (N=32 cells). (D) Univariate scatter plot of the number of bundles in RPE1 cells treated with MG-132 counted in the end-on view of the spindle in control cells (gray) and HAUS6-depleted cells (yellow). N=32 control cells and N=39 HAUS6-depleted cells. (E) The PRC1-GFP intensity profiles in RPE1 cells treated with MG-132 measured in the end-on view of the spindle in control cells (gray) and after HAUS6 depletion (yellow). The blue line in the inset marks the measured region (width: 2.5 µm). Mean (thick lines) and SEM (shaded areas). (F) Univariate scatter plot of overlap length divided by spindle length in RPE1 cells treated with MG-132 measured in the side view of the spindle in control cells (gray) and HAUS6-depleted cells (yellow). N=75 bundles in 32 control cells and N=74 bundles from 39 HAUS6-depleted cells. (D and F) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Statistical analysis (C, D and F) Mann–Whitney U test; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). All images are adjusted for clarity so that all PRC1 bundles are visible in each cell (see Materials and methods). Scale bars, 2 µm.
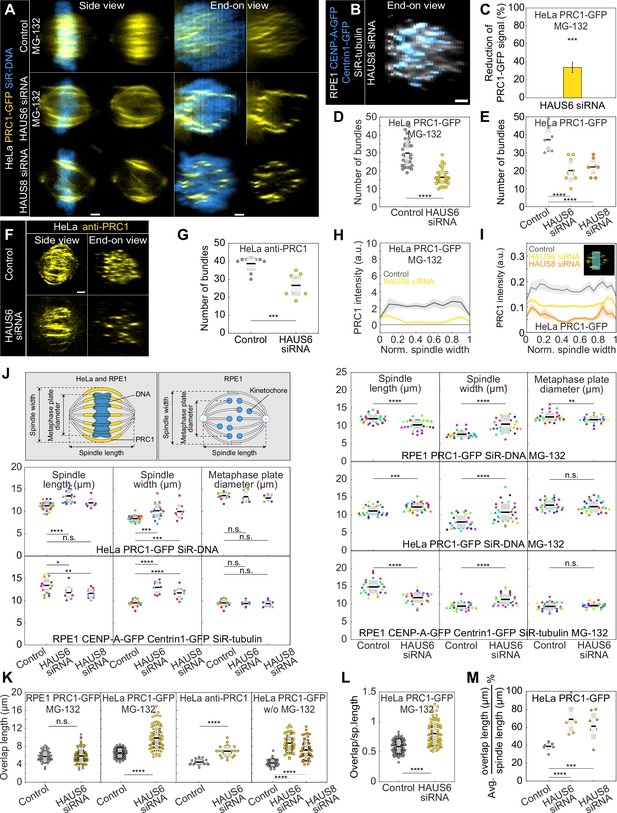
Spindles without augmin are wider with fewer overlaps, which occupy a larger portion of the spindle length and accumulate at the spindle periphery.
(A) Four columns represent live images of metaphase spindles in HeLa cells stably expressing PRC1-GFP (yellow) and stained with SiR-DNA (blue) in control (top) and after HAUS6 (middle) or HAUS8 (bottom) depletion. Control and HAUS6-depleted cells are treated with MG-132. 1st and 2nd column: side view of the spindle; 3rd and 4th column: end-on view of the same spindle, showing a barrel-like arrangement of PRC1-labeled bundles after augmin depletion. Side views are sum projections of 5 central z-slices (∆z=0.5 µm) and end-on views are sum projections of 10 central z-slices (∆z=0.083 µm). (B) End on view of RPE1 cell stably expressing CENP-A-GFP and Centrin1-GFP (both in blue) and stained with SiR-tubulin (gray) after HAUS8 depletion. End-on view is a sum projection of 10 central z-slices (∆z=0.083 µm). (C) Reduction of the PRC1 signal in HeLa cells stably expressing PRC1-GFP and treated with MG-132, measured in sum intensity projection of 10 central z-slices following HAUS6 depletion. Values are shown as mean ± SEM. P-values were calculated using the absolute values of the PRC1 signal intensity following HAUS6 depletion (N=39 cells), compared to the absolute values of the PRC1 signal intensity in control cells (N=33 cells). (D) Univariate scatter plot of the number of PRC1-labeled bundles in HeLa cells stably expressing PRC1-GFP treated with MG-132, counted in the end-on view of the spindle in control (gray) and HAUS6-depleted cells (yellow). N=32 control and 39 HAUS6-depleted cells. (E) Univariate scatter plot of the number of PRC1-labeled bundles in HeLa cells stably expressing PRC1-GFP, counted in the end-on view of the spindle in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) cells without MG-132 treatment. N=10 cells for all conditions. (F) Fixed control and HAUS6 siRNA-treated HeLa cells immunostained for PRC1 (yellow). Images are sum intensity projections of 5 central z-planes in a side view and sum projection of 10 central z-planes in an end-on view. The corresponding number of PRC1-labeled bundles (G) is shown in an univariate scatter plot. N=10 control and 10 HAUS6-depleted cells. (H) The PRC1-GFP intensity profile in HeLa cells stably expressing PRC1-GFP and treated with MG-132, measured in the end-on view of the spindle in control cells (gray) and after HAUS6 (yellow) depletion. The blue line in the inset marks the measured region (width: 2.5 µm). Mean (thick lines) and SEM (shaded areas). (I) Same as (H) but cells are not treated with MG-132 and HAUS8 depletion (orange) is included in the plot. (J) Schematic representations of spindle geometry measurements in RPE1 and HeLa cells stably expressing PRC1-GFP (yellow) and stained with SiR-DNA (blue) and RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (both in yellow) and stained with SiR-tubulin (white). Univariate scatter plots of the spindle length (left), width (middle) and diameter of the metaphase plate (right) are shown in this order on both left and right panels. The cell type from which the measurement was obtained is indicated in each panel. The measurements were taken in control, HAUS6- or HAUS8-depleted cells. Left panels show cells treated with MG-132 and right panels represent untreated cells. Each color in the plot corresponds to data obtained in one cell. N=36 control cells and 33 HAUS6-depleted cells following MG-132 treatment of the RPE1 PRC1-GFP cell line; N=33 control cells and 38 HAUS6-depleted cells following MG-132 treatment of the HeLa PRC1-GFP cell line; N=31 control cells and 31 HAUS6-depleted cells following MG-132 treatment of the RPE1 CENP-A-GFP Centrin1-GFP cell line; N=10 control, HAUS6- and HAUS8-depleted HeLa PRC1-GFP and RPE1 CENP-A-GFP Centrin1-GFP cells. (K) Overlap length measured in the side view of the spindle of control (gray) and HAUS6-depleted (yellow) RPE1 cells stably expressing PRC1-GFP and treated with MG-132 (left). N=72 overlaps in 32 control cells and 74 overlaps in 39 HAUS6-depleted cells. Overlap length measured in the side view of the spindle of control (gray) and HAUS6-depleted (yellow) HeLa cells stably expressing PRC1-GFP and treated with MG-132 (middle, left). N=96 overlaps in 33 control cells and 90 overlaps in 39 HAUS6-depleted cells. Overlap length measured in the side view of the spindle of control (gray) and HAUS6-depleted (yellow) cells immunostained for PRC1 without MG-132 treatment (middle, right). N=27 overlaps in 10 control cells and 23 overlaps in 10 HAUS6-depleted cells. Overlap length measured in the side view of the spindle of control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) HeLa cells stably expressing PRC1-GFP without MG-132 treatment (right). N=50 overlaps in 10 cells for each condition. Each color in the plot corresponds to data obtained from one cell. (L) Univariate scatter plot of an overlap length divided by spindle length in control (gray) and HAUS6-depleted (yellow) HeLa cells stably expressing PRC1-GFP and treated with MG-132. N=96 overlaps in 33 control cells and 90 overlaps in 39 HAUS6-depleted cells. (M) Univariate scatter plot of an average overlap length divided by spindle length in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) HeLa cells stably expressing PRC1-GFP. N=10 overlaps in 10 cells for each condition. (D, E, G, J–M) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. All images are adjusted for clarity (see Materials and methods). Statistical analysis (C, G and L) Mann-Whitney U test; (E, J left panels, M) ANOVA with post-hoc Tukey test; (J and K) t-test for samples that followed normal distribution or Mann–Whitney U test for samples that significantly departured from normality, determined using the Shapiro-Wilk test; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). Scale bars, 2 µm.
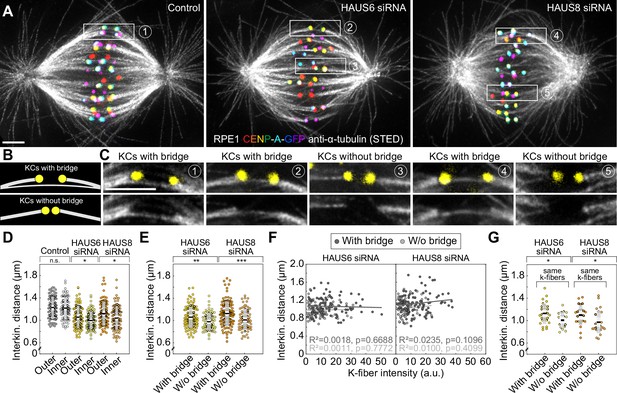
The reduction of the interkinetochore distance after augmin depletion is related to the impairment of bridging fibers.
(A) STED superresolution images of microtubules immunostained for α-tubulin (gray) in control (left), HAUS6- (middle) and HAUS-8 depleted (right) RPE1 cells stably expressing CENP-A-GFP (rainbow, confocal). Images are maximum intensity projections and kinetochores are color-coded for depth from blue to red with the Spectrum LUT in ImageJ. (B) The schematic representation of a kinetochore pair (KCs) with (top) and without (bottom) bridging fiber (See Results). (C) Enlarged boxes show KCs with or without a bridging fiber in control (left), HAUS6- (middle), and HAUS8- (right) depleted RPE1 cells. Images represent single z-plane taken from spindles in (A) and smoothed with 0.75-mm-sigma Gaussian blur. Kinetochores are shown in yellow. (D) Univariate scatter plot of the interkinetochore distance in control (gray), HAUS6- (yellow), and HAUS8- (orange) depleted cells with kinetochore pairs divided based on their distance from the long (pole-to-pole) spindle axis (outer kinetochore pairs shown in darker colors and inner in lighter colors). N=30 cells in all three conditions; 78 and 80 outer and inner kinetochore pairs for control, respectively; 84 and 96 outer and inner kinetochore pairs for HAUS6 depletion, respectively; 88 and 92 outer and inner kinetochore pairs for HAUS8 depletion, respectively. (E) Univariate scatter plot of the interkinetochore distance in HAUS6- (yellow) and HAUS8- (orange) depleted cells. Kinetochore pairs are divided into two groups: with bridging fiber (darker colors) and without bridging fiber (lighter colors). N=30 cells in HAUS6/8-depleted cells; 106 pairs with and 74 kinetochore pairs without bridging fibers in cells following HAUS6 depletion, respectively; 110 and 70 kinetochore pairs with and without bridging fibers in cells following HAUS8 depletion, respectively. (F) The correlation of the interkinetochore distance and the k-fiber intensity for kinetochore pairs with (dark gray) and without (light gray) bridging fiber in HAUS6- (left) and HAUS8-depleted (right) cells. (G) Univariate scatter plot of the interkinetochore distance for HAUS6- (yellow) and HAUS8-depleted (orange) cells. Kinetochore pairs are divided into two groups: with bridging fiber (darker colors) and without bridging fiber (lighter colors), but in this case both groups have the same k-fiber intensity. N=27 kinetochore pairs with a bridging fiber and 18 kinetochore pairs without a bridging fiber in HAUS6-depleted cells, respectively; N=23 kinetochore pairs with and N=25 kinetochore pairs without a bridging fiber in HAUS8-depleted cells. (D, E and G) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Statistical analysis (D, E and G) t-test for samples that followed normal distribution or Mann–Whitney U test for samples that significantly departured from normality, determined using the Shapiro-Wilk test; (F) linear regression; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns). All images are adjusted for clarity based on the intensity of astral microtubules in each image (see Materials and methods). Scale bars, 2 µm.
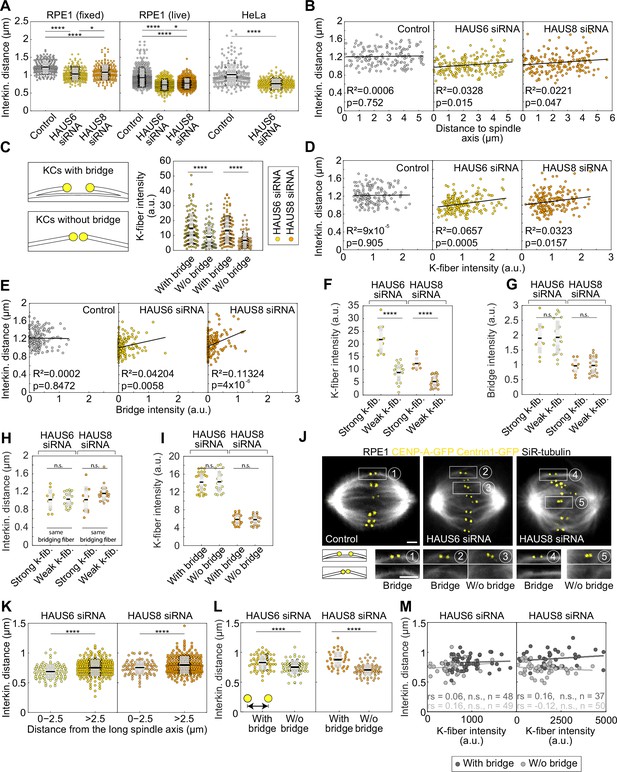
Augmin-generated bridging fibers have a distinct role in the regulation of kinetochore tension.
(A) Univariate scatter plot of the interkinetochore distance in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin (left). N=158 kinetochore pairs in 30 control cells and 180 kinetochore pairs in 30 HAUS6- and HAUS8-depleted cells. Univariate scatter plot of the interkinetochore distance in control (gray), HAUS6- (yellow), and HAUS8-depleted (orange) RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP and stained with SiR-tubulin (middle). N=446 kinetochore pairs in 10 control cells, 356 kinetochore pairs in 10 HAUS6-depleted cells, and 374 kinetochore pairs in 30 HAUS8-depleted cells. Univariate scatter plot of the interkinetochore distance in control (gray) and HAUS6-depleted (yellow) HeLa cells stably expressing CENP-A-GFP and Centrin1-GFP and stained with SiR-tubulin (right). N = >100 kinetochore pairs in 5 control and HAUS6-depleted cells. (B) Correlation of the interkinetochore distance and distance to spindle axis in control (gray), HAUS6- (yellow) and HAUS8-depleted (orange) RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. (C) Univariate scatter plots of the k-fiber intensity for kinetochore pairs with bridging fiber (dark colors) and without bridging fiber (light colors) in HAUS6- (yellow) and HAUS8-depleted (orange) RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. Schematic representation of a kinetochore pair (KCs) with and without bridging fiber (See Results). N=30 HAUS6-depleted cells with 106 kinetochore pairs with bridging fibers and 74 kinetochore pairs without bridging fibers. N=30 HAUS8-depleted cells with 110 kinetochore pairs with bridging fibers and 70 kinetochore pairs without bridging fibers. (D) Correlation of the interkinetochore distance and k-fiber intensity for kinetochore pairs in control (gray), HAUS6- (yellow) and HAUS8- (orange) depleted RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. (E) Correlation of the interkinetochore distance and bridging fiber intensity for kinetochore pairs in control (gray), HAUS6- (yellow) and HAUS8- (orange) depleted RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. (B, D and E) N=158, 180 and 180 kinetochore pairs in 30 control, HAUS6- and HAUS8-depleted cells, respectively. For D and E, bridging and k-fiber intensities were normalized to the average intensity of two outermost k-fibers in each cell and negative intensities were set to a value zero. (F) Univariate scatter plots of the k-fiber intensity for kinetochore pairs with stronger k-fiber (dark colors) and with weaker k-fiber (light colors) within subgroups of kinetochore pairs with or without bridging fiber after HAUS6 (yellow) or HAUS8 (orange) depletion in RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. (G) Univariate scatter plots of the bridging fiber intensity of kinetochore pairs in (F). (H) Univariate scatter plots of the interkinetochore distance for kinetochore pairs in (F). (F–I) N=11 kinetochore pairs with stronger k-fiber and 20 kinetochore pairs with weaker k-fiber in 30 HAUS6-depleted cells. N=10 kinetochore pairs with stronger k-fiber and 23 kinetochore pairs with weaker k-fiber in 30 HAUS8-depleted cells. (I) Univariate scatter plot of the k-fiber intensity for kinetochore pairs with bridging fibers (dark colors) and without bridging fibers (light colors), after HAUS6 (yellow) or HAUS8 (orange) depletion in RPE1 cells stably expressing CENP-A-GFP and immunostained for α-tubulin. K-fiber intensities are chosen to be similar within the subgroups with or without a bridging fiber to exclude the contribution of k-fibers. N=27 kinetochore pairs with a bridging fiber and 18 kinetochore pairs without a bridging fiber in HAUS6-depleted cells. N=23 kinetochore pairs with a bridging fiber and 25 kinetochore pairs without a bridging fiber in HAUS8-depleted cells. (J) Live images (single z-plane) of metaphase spindles in RPE1 cells stably expressing CENP-A-GFP and Centrin1-GFP (both in yellow) and stained with SiR-tubulin (gray). Enlarged boxes show kinetochore (KC) pairs with or without a bridging fiber. All images are adjusted for clarity (see Materials and methods). Scale bars, 2 µm. (K) Univariate scatter plot of the interkinetochore distance in HAUS6- (yellow) or HAUS8- (orange) depleted cells depending on the 3D-distance from the long spindle axis. N=10 HAUS6-depleted cells with 97 and 259 kinetochore pairs in the intervals of 0–2.5 and >2.5, respectively. N=10 HAUS8-depleted cells with 95 and 265 kinetochore pairs in the intervals of 0–2.5 and >2.5, respectively. (L) Univariate scatter plot of the interkinetochore distance in HAUS6- (yellow) and HAUS8- (orange) depleted cells. N=10 cells with >50 kinetochore pairs. Kinetochore pairs are divided into two groups: with bridging fiber (dark gray) and without bridging fiber (light gray). N=65 kinetochore pairs with and without a bridging fiber in 10 HAUS6-depleted cells; N=42 kinetochore pairs with a bridging fiber and 74 kinetochore pairs without a bridging fiber in HAUS8-depleted cells. (M) Correlation of the interkinetochore distance and k-fiber intensity for kinetochore pairs with (dark gray) and without (light gray) a bridging fiber in HAUS6 siRNA (left) and HAUS8 siRNA (right)-treated cells. The data represent a subset from L, in which only kinetochore pairs with k-fibers well isolated from neighboring microtubules were taken into account. The interkinetochore distance values for kinetochore pairs with and without bridging fibers were 0.83±0.02 and 0.76±0.02 µm for HAUS6 siRNA- and 0.89±0.02 and 0.73±0.01 µm for HAUS8 siRNA-treated cells, respectively, in agreement with results from panel L. rs, Spearman correlation coefficient. (A, C, F–I, K, L) Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray) and mean value (black). All results were obtained from three independent experiments. Statistical analysis (A left and middle) ANOVA with post-hoc Tukey test; (A right) t-test; (B, D, E and M) linear regression; (C, F–I, K and L) t-test for samples that followed normal distribution or Mann–Whitney U test for samples that significantly departured from normality, determined using the Shapiro-Wilk test; p-value legend:<0.0001 (****), 0.0001–0.001 (***), 0.001–0.01 (**), 0.01–0.05 (*), ≥0.05 (ns).
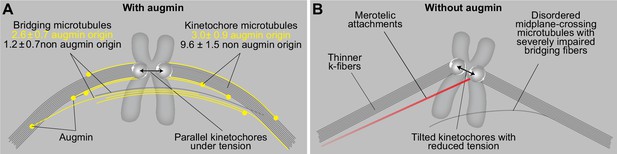
A model of augmin-dependent nucleation of bridging microtubules with their contribution to mitotic fidelity.
(A) Bridging microtubules are to a large extent formed by augmin-dependent nucleation. They ensure the alignment of sister kinetochores parallel to the spindle axis and the interkinetochore tension. Augmin-nucleated microtubules (yellow) and microtubules nucleated through other pathways (gray) in bridging and k-fibers are shown together with the number of microtubules in each group, as estimated from HAUS6 depletion experiments (See Results). (B) Impaired structure of bridging fibers upon augmin depletion leads to weaker interkinetochore tension and increased tilt of the kinetochores, which puts kinetochores at risk of interacting with additional microtubules (red), resulting in merotelic attachments.
Videos
RPE1 cell stably expressing CENP-A-GFP and Centrin1-GFP (16-colors) following Mad2 depletion.
Kinetochores are color-coded for depth from blue to red with the 16 Colors LUT and noise was processed with the Despeckle function in ImageJ. Scale bar, 2 µm.
RPE1 cell stably expressing CENP-A-GFP and Centrin1-GFP (16-colors) following Mad2/HAUS6 codepletion.
Kinetochores are color-coded for depth from blue to red with the 16 Colors LUT and noise was processed with the Despeckle function in ImageJ. Scale bar, 2 µm.





