Syntaxin-6 delays prion protein fibril formation and prolongs the presence of toxic aggregation intermediates
Figures
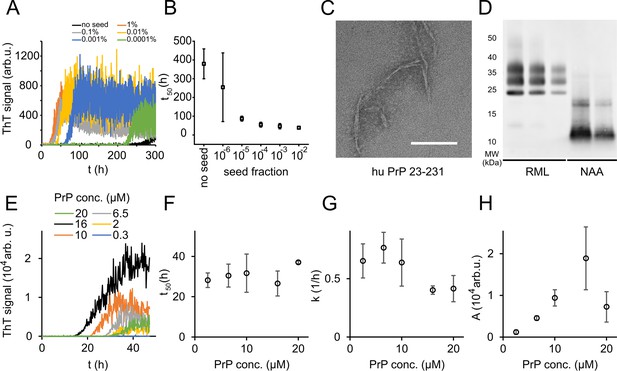
Native aggregation assay (NAA) of human PrP (23-231).
(A) Seed titration experiment using seed generated from hPrP23 fibrils formed de novo in NAA, seed fraction (w/w), 2.5 µM hPrP23, pH 6.8, 42°C. (B) Plot of lag phase vs. fraction of seed, mean ± SD, n = 3. (C) TEM image of hPrP23 fibrils formed after 166 hr incubation. (D) Western blot (ICSM35) of ex vivo RML prions and NAA aggregation endpoint samples after digestion with proteinase K (50 µg/ml, 30 min) and NaPTA precipitation. (E–H) Titration of PrP concentration (0.3–20 µM) in ThT aggregation assays containing 0.01% seed, pH 6.8, 42°C. (E) Plot of ThT fluorescence vs. time. (F) Plot of lag time t50 vs. PrP concentration. (G) Plot of elongation rate constant k vs. PrP concentration. (H) Plot of fluorescence amplitude A vs. PrP concentration. Source data is available at https://doi.org/10.17632/yggpkrgnx8.1.
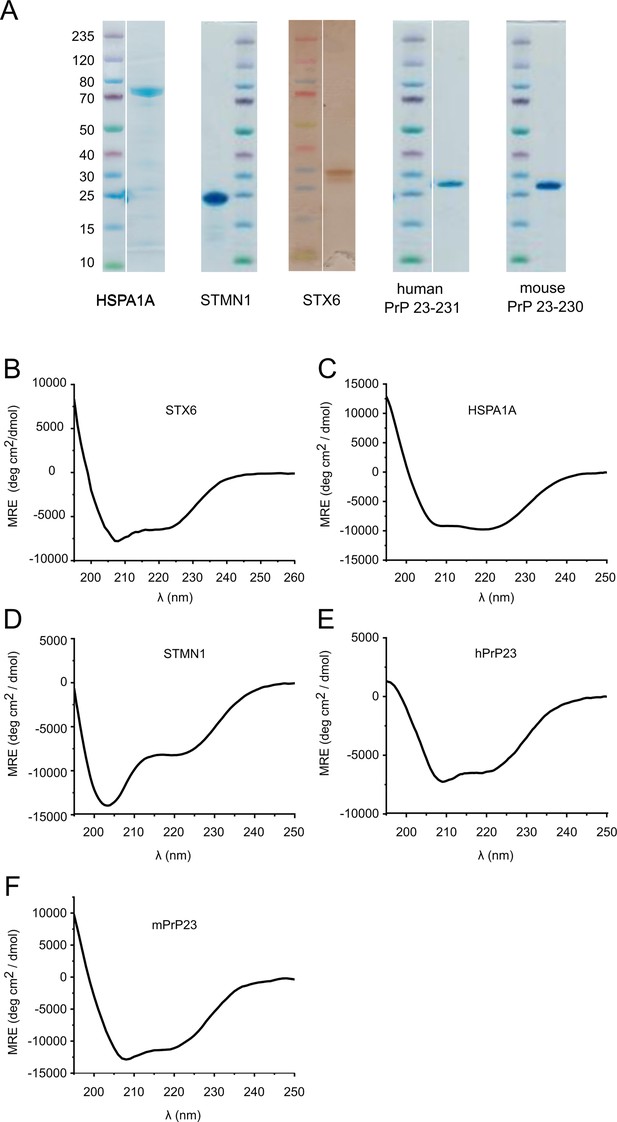
Protein characterization.
(A) Coomassie/silver stained gels of purified hPrP23, mPrP23, and candidate proteins. (B) Circular dichroism spectrum of purified syntaxin-6 (STX6) protein showing a mostly alpha helical shape with minima at 208 nm and 223 nm. (C–F) Circular dichroism spectra of Hsp70 (HSPA1A), stathmin 1 (STMN1), human PrP 23-231,129M (hPrP23), and mouse PrP 23-231 (mPrP23). Source data is available at https://doi.org/10.17632/yggpkrgnx8.1.

Optimization of native aggregation assay (NAA).
(A) Plot of mPrP23 ThT fluorescence over time at 37, 42, and 50°C (pH 7.4). (B) mPrP23 NAA aggregation kinetics at pH 6.6, 6.8, 7.0, and 7.2 at 42°C. (C) NAA aggregation kinetics of hPrP23 (2.5 µM) with Zr vs. glass beads, pH 6.8 at 42°C. (D–F) mPrP23 seed titration experiments comparing seeding from synthetic PrP fibrils (Sp) and purified prion rods from mouse prion strains ME7 and RML. (D) Plot of time to half-maximal fluorescence (t50) (E) elongation rate constant (k), and (F) ThT amplitudes as functions of seed concentration; mean ± SD, n = 5. (G) TEM image of mPrP23 fibrils formed after 47 hr incubation at pH 6.8, 42°C; scale bar 100 nm. (H) Silver stain gel of hPrP23 monomer and NAA product after 30 min PK digestion (50 µg/ml). Source data is available at https://doi.org/10.17632/yggpkrgnx8.1.
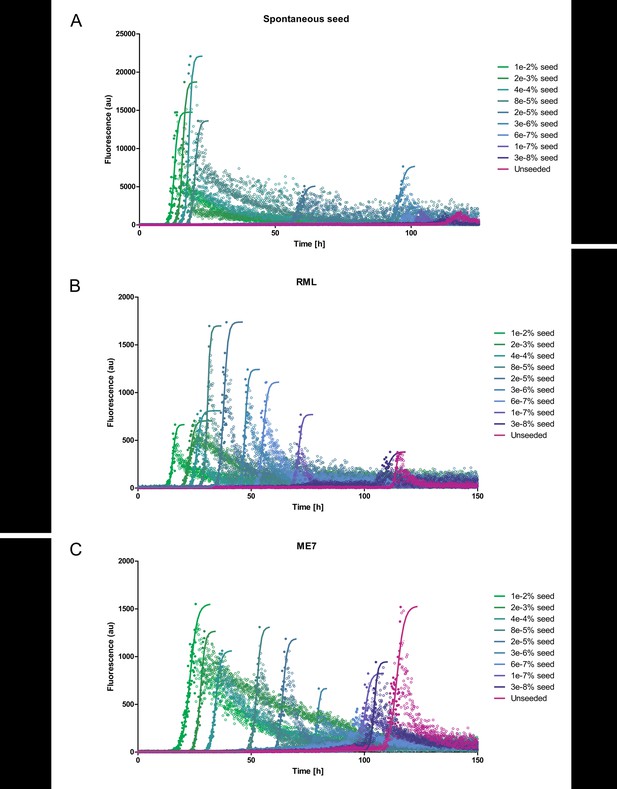
mPrP23 seeding kinetics.
Graphs of native aggregation assay (NAA) on mPrP23 (10 µM) seed titration experiments demonstrate seed concentration-dependent acceleration of fibril formation. (A) First-generation (spontaneous) seed. (B) Seed generated from RML mouse strain. (C) Seed generated from ME7 mouse strain. Solid lines indicate sigmoidal fits of the data, n = 3. Post aggregation decay of ThT fluorescence, which was likely due to adsorption and sedimentation, was not included in the fits.
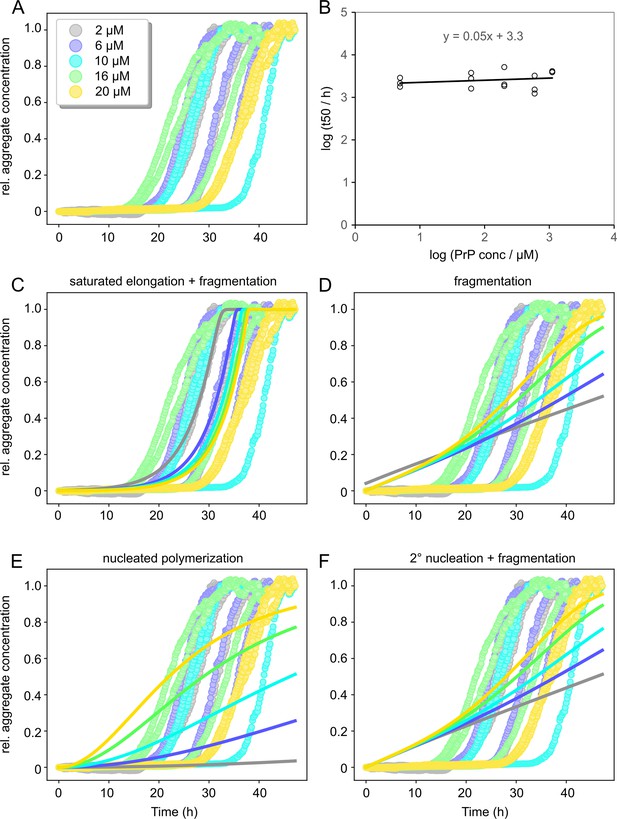
Amylofit analysis of concentration-dependent native aggregation assay (NAA) data.
Aggregation data from experiment shown in Figure 1E were smoothed by moving average and normalized (A) and then analyzed in the Amylofit framework to generate t50 values (B; n = 3). (C–F) Global fitting of data using different models for aggregation kinetics as indicated. Only the model including saturated elongation adequately fitted the data.
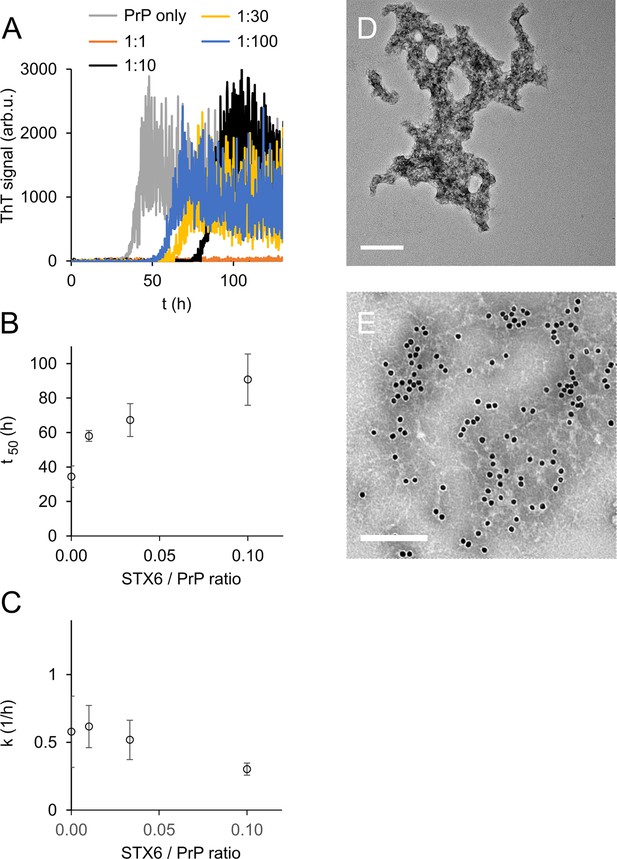
Syntaxin-6 (STX6) delays hPrP23 fibril formation at molar ratios of 1:1–1:100; 2.5 µM hPrP23, pH 6.8, 42°C, 0.01% seed.
(A) ThT fluorescence vs. time. (B) Plot of lag phase t50 vs. molar ratio of STX6/PrP; mean ± SD, n = 3. (C) Plot of elongation rate constant k vs. molar ratio of syntaxin-6/PrP. (D) TEM image of hPrP23 co-aggregated with syntaxin-6 at 1:10 (STX6/PrP) molar ratio under standard native aggregation assay (NAA) conditions for 116 hr. (E) Immuno-TEM image of hPrP23 – syntaxin-6 co-aggregate cluster after 100 hr aggregation. Syntaxin-6 is labeled with anti-syntaxin-6 Ab/10 nm anti-rabbit immunogold beads; scale bars 200 nm.
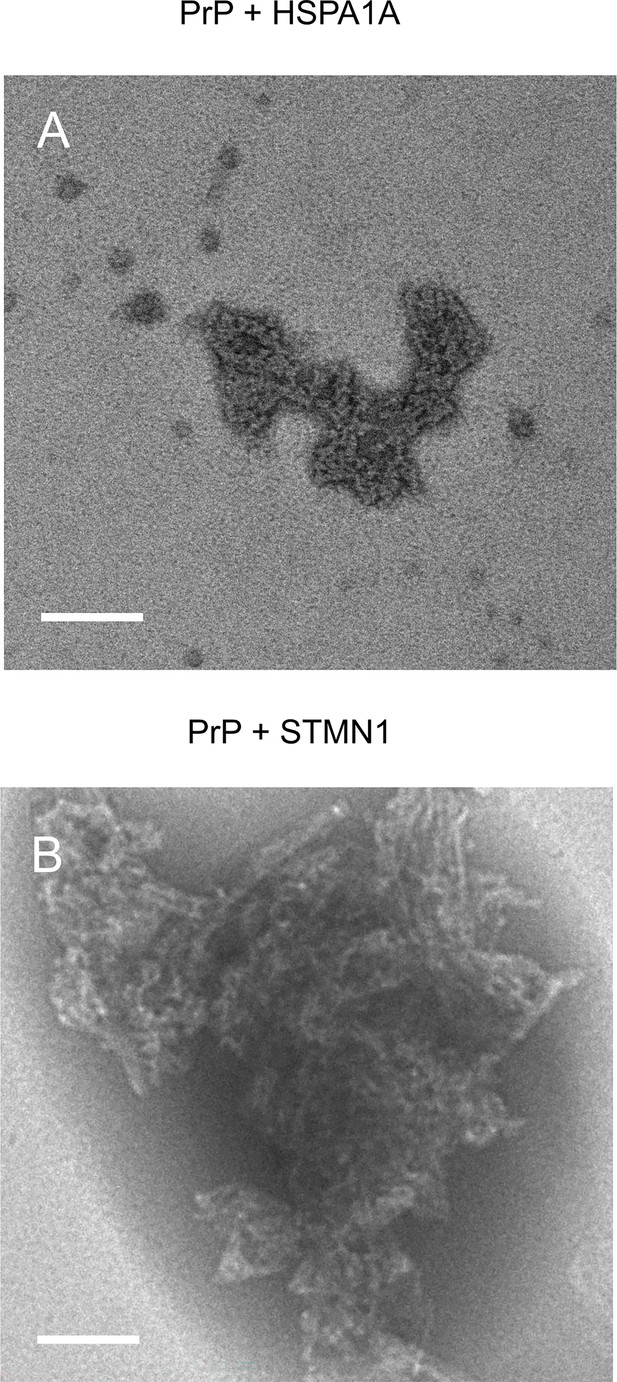
TEM images of native aggregation assay (NAA) aggregation endpoints of hPrP23 co-aggregated with HSP70 (HSPA1A) or stathmin 1 (STMN1).
(A) PrP forms amorphous aggregates in the presence of HSPA1A at 1:10 molar ratio. (B) PrP forms fibrillar aggregates in the presence of STMN1 at 1:10 molar ratio; scale bar 100 nm.
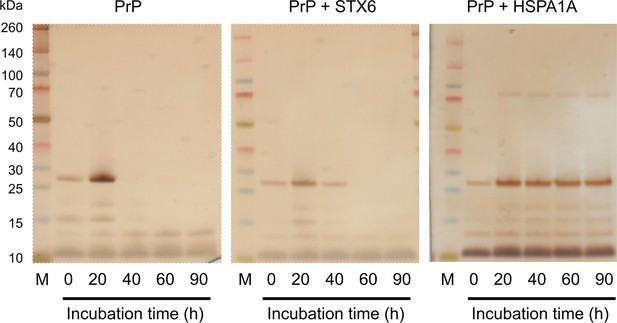
Sedimentation assay of hPrP23.
hPrP23 was incubated for 0–90 hr alone or co-aggregated with syntaxin-6 (1:10 molar ratio) or HSPA1A (1:10 molar ratio). Protein samples were centrifuged; supernatants were collected, and analyzed by silver-stained SDS-PAGE. Source data is available at https://doi.org/10.17632/yggpkrgnx8.1.
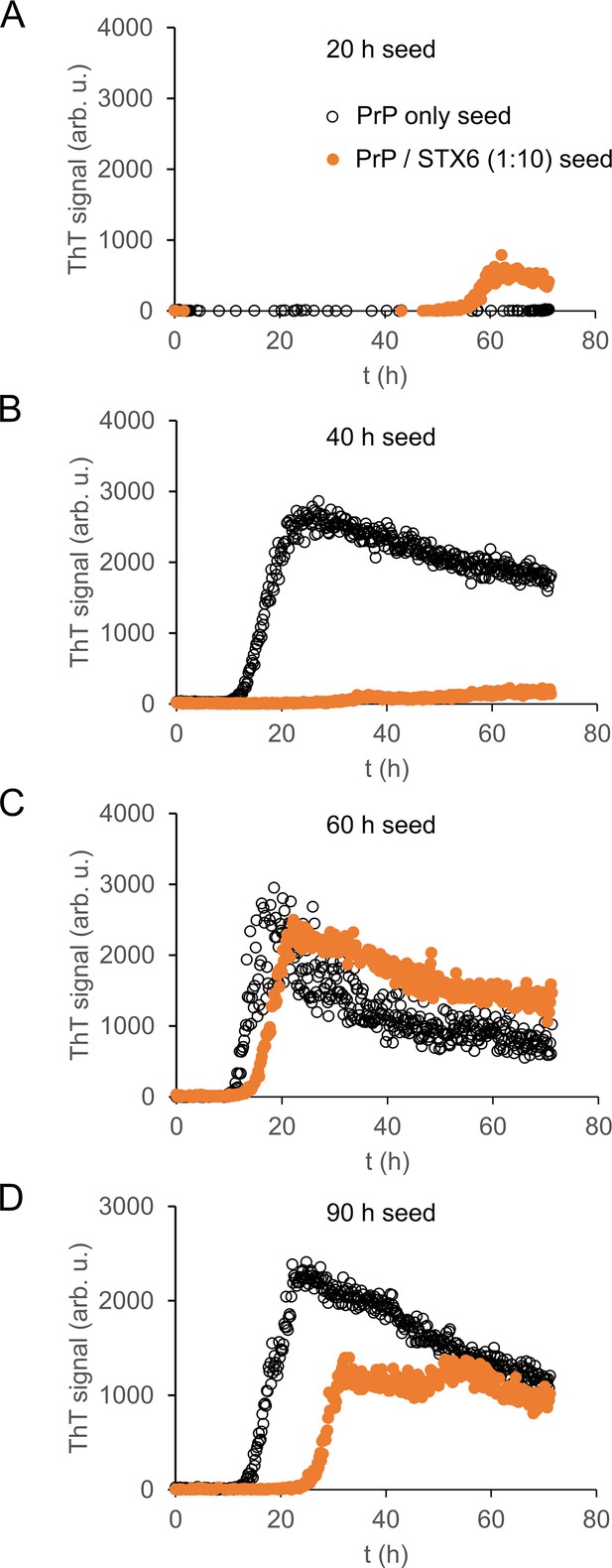
Seeding capacity of aggregation intermediates.
Aggregates of hPrP23 (black) or hPrP23 co-aggregated with syntaxin-6 (STX6, 1:10 molar ratio; orange) collected at 20 hr (A), 40 hr (B), 60 hr (C), and 90 hr (D) were used to seed a second aggregation assay using hPrP23 as substrate; seed concentration 0.1% (w/w). Aggregation time points correspond to those used in toxicity assays. Graphs represent averaged data of three replicate experiments.
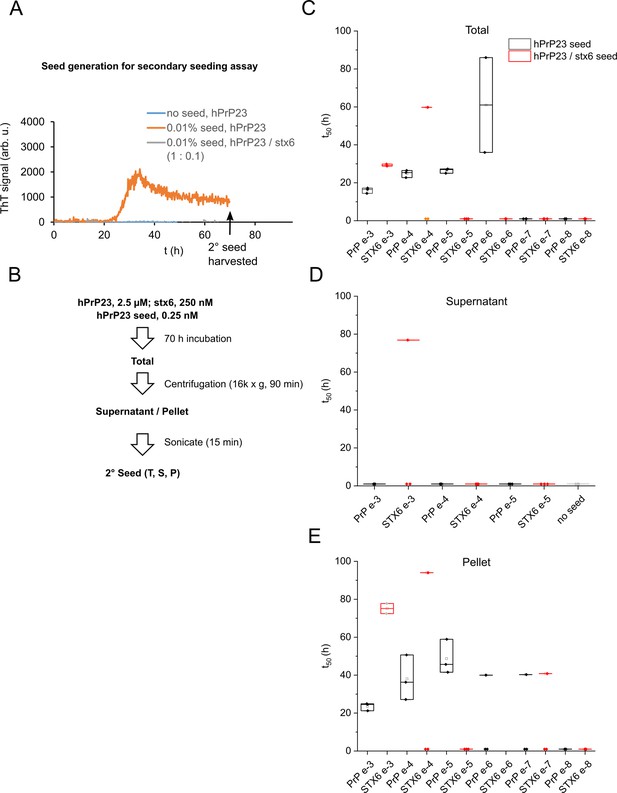
Secondary seeding seed dilution experiment of hPrP23 aggregates.
(A) Native aggregation assay (NAA) experiment of hPrP23 (2.5 µM) in the absence (orange) or presence of stx6 (0.25 µM, gray). (B) Endpoint aggregates (t = 70 hr) were separated into total, soluble (supernatant), and insoluble (pellet) fractions, which were resuspended to the original volume and sonicated to generate secondary seeds. (C–E) Box plots of times to half maximal fluorescence (t50) in secondary hPrP23 seeding assays seeded at 10–3 to 10–8 seed to monomer ratio using total, supernatant, and pellet seed fractions; n = 3, data points at the bottom of graphs indicate samples that failed to aggregate.
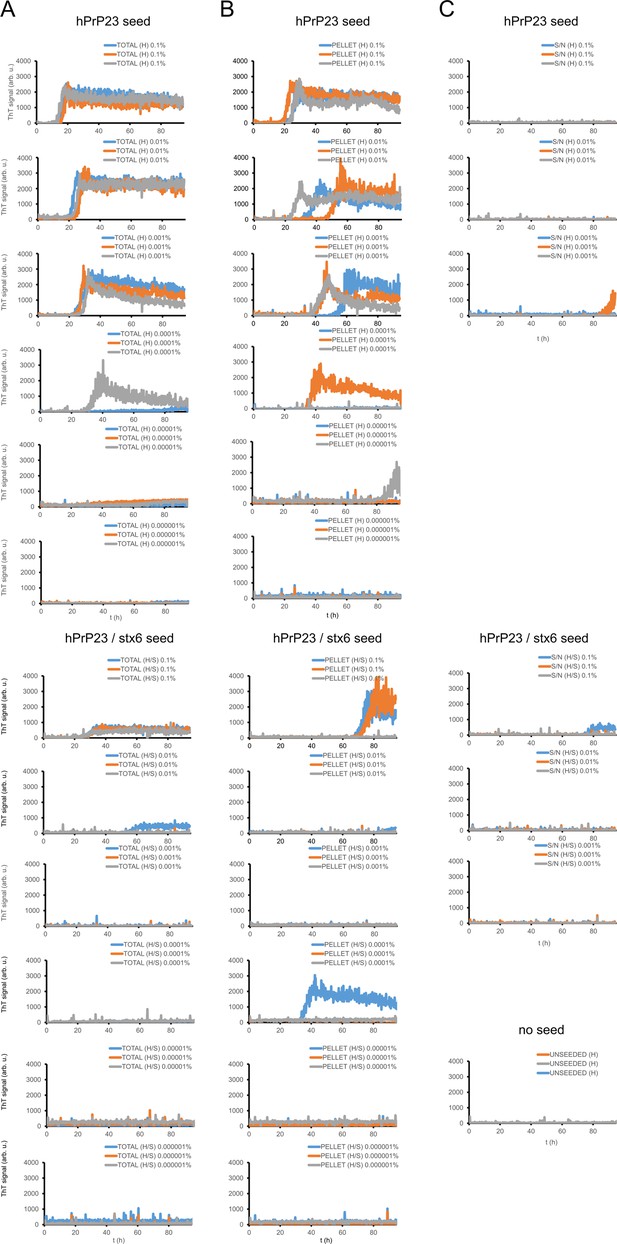
Secondary seeding seed dilution kinetics.
Kinetic traces of hPrP23 native aggregation assay (NAA) secondary seeding assays at 10–3 to 10–8 seed to monomer ratio described in Figure 2—figure supplement 4: (A) total, (B) pellet, and (C) supernatant seed fractions and unseeded kinetics; n = 3, only dilutions 10–3 to 10–5 are plotted for supernatant seeds.
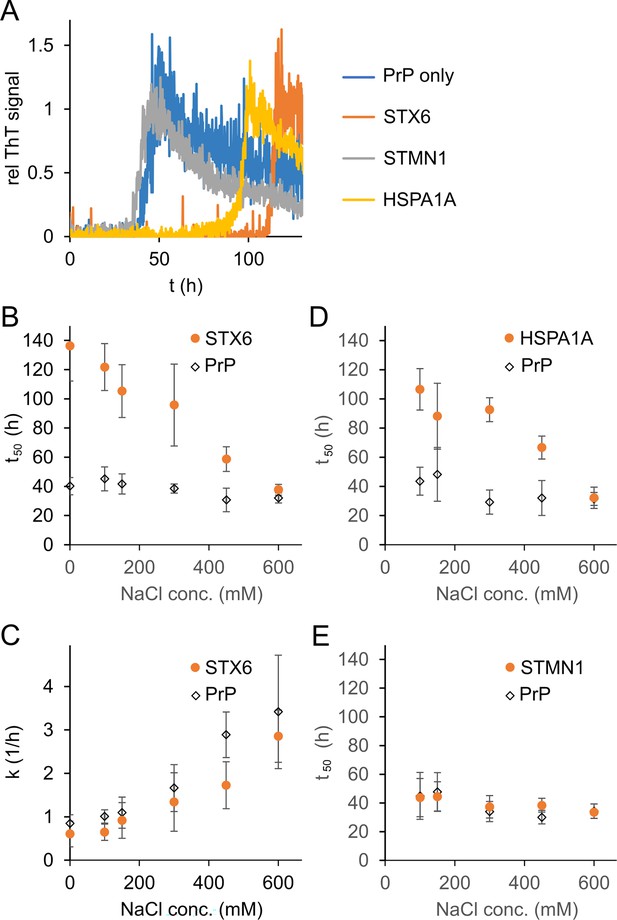
Salt-dependent inhibition of fibril formation.
Syntaxin-6, STMN1, HSPA1A 1:10 molar ratio were incubated with hPrP23. (A) Plot of relative ThT fluorescence profiles vs. time in NAA (150 mM NaCl, 42 °C, 0.01% seed,). (B) Plot of lag phase t50 vs. NaCl concentration for hPrP23 alone vs. co-aggregated with syntaxin-6 (STX6) at 1:10 molar ratio. (C) Plot of elongation rate constant k vs. NaCl concentration. (D) Plot of lag phase t50 vs. NaCl concentration for hPrP23 alone vs. co-aggregated with HSPA1A at 1:10 molar ratio. (E) Plot of lag phase t50 vs. NaCl concentration for hPrP 23–231 alone vs. co-aggregated with STMN1 at 1:10 molar ratio; mean ± SD, n = 3.
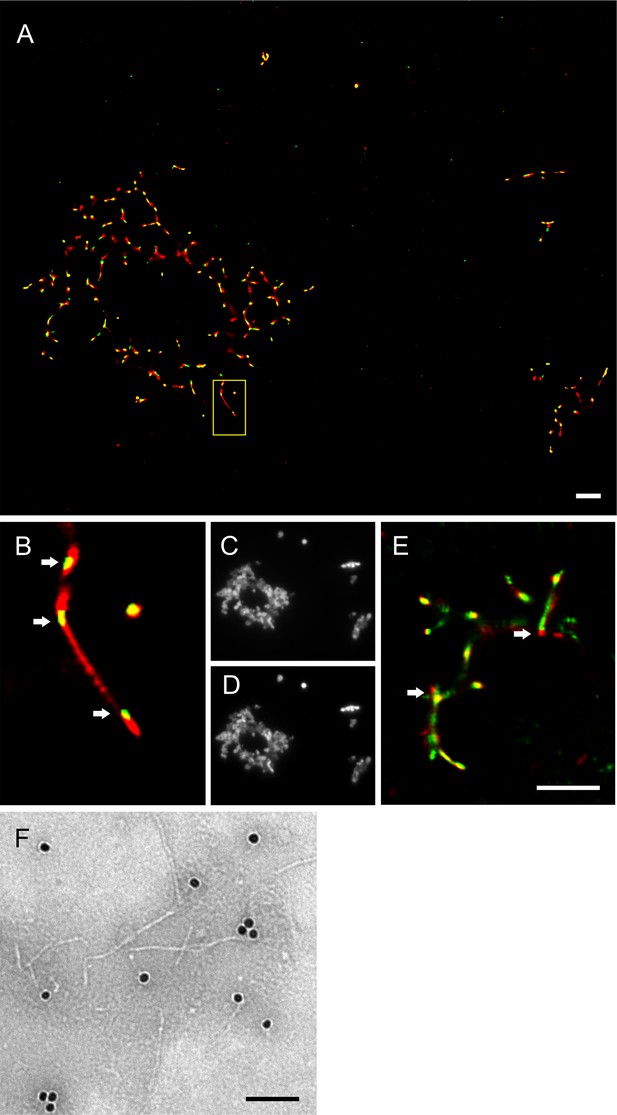
Imaging of hPrP23 fibrils incubated with syntaxin-6.
hPrP23 (2.5 µM) was pre-aggregated in native aggregation assay (NAA) for 115 hr and incubated with syntaxin-6 (250 nM) for 1 hr. hPrP was visualized by TAB imaging using 10 nM Nile red dye; syntaxin-6 was labeled with AlexaFluor488 (A–D) or AlexaFluor647 (E) and imaged by dSTORM. (A) SR image overlay shows syntaxin-6 binding at hotspots and PrP fibril ends. (B) Magnified area from (A). (C) Widefield image taken with green laser (561 nm) illumination. (D) Widefield image taken under blue laser (473 nm) illumination. (E) SR image overlay of PrP and syntaxin-6-AF647 images; scale bars 2 µm. (F) TEM Immuno-gold staining of hPrP23 fibrils incubated with syntaxin-6; scale bar 100 nm.
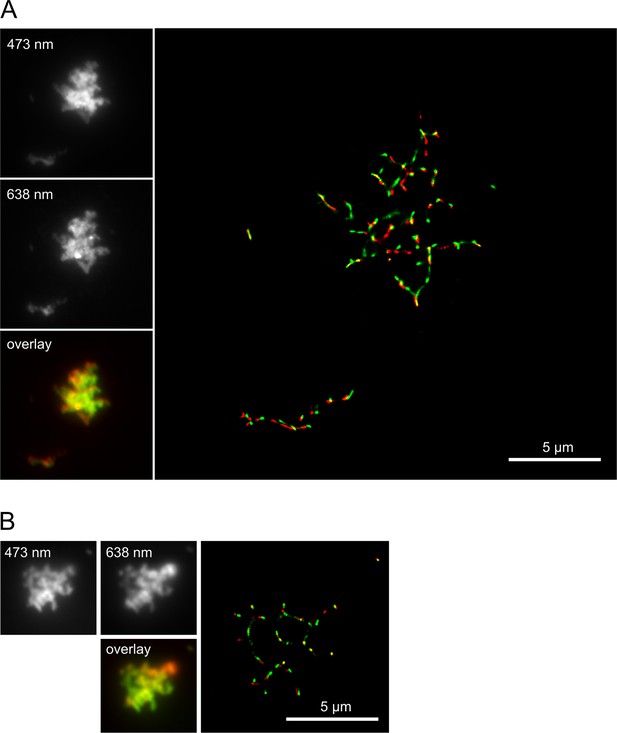
Overlay of dSTORM SR images taken at 473 nm and 638 nm excitation.
Co-aggregates of hPrP23 and syntaxin-6 formed in native aggregation assay (NAA) under conditions of Figure 3. (A) Co-aggregation of hPrP23 labeled with AlexaFluor 647-NHS (2.5 µM, 3% labeled, 95% unlabeled) with 10 nM syntaxin-6 labeled with AlexaFluor488-NHS. (B) Co-aggregation of PrP labeled with AlexaFluor 488-NHS (2.5 µM, 5% labeled, 95% unlabeled) with syntaxin-6-AlexaFluor647 at 1:10 molar ratio (syntaxin-6, 250 nM, 4% labeled, 96% unlabeled). Insets: widefield images with 473 nm and 638 nm excitation and composite image of both channels.
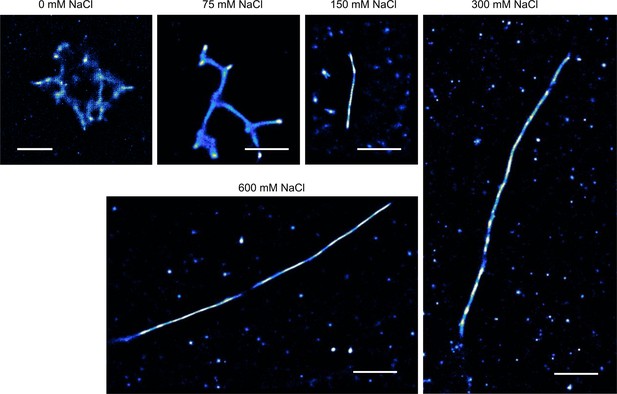
TAB SR microscopy images of aggregation endpoints of hPrP23.
hPrP23 was aggregated in NAA with increasing salt concentrations from 0 mM to 600 mM NaCl for 140 hr; scale bars 2 µm.
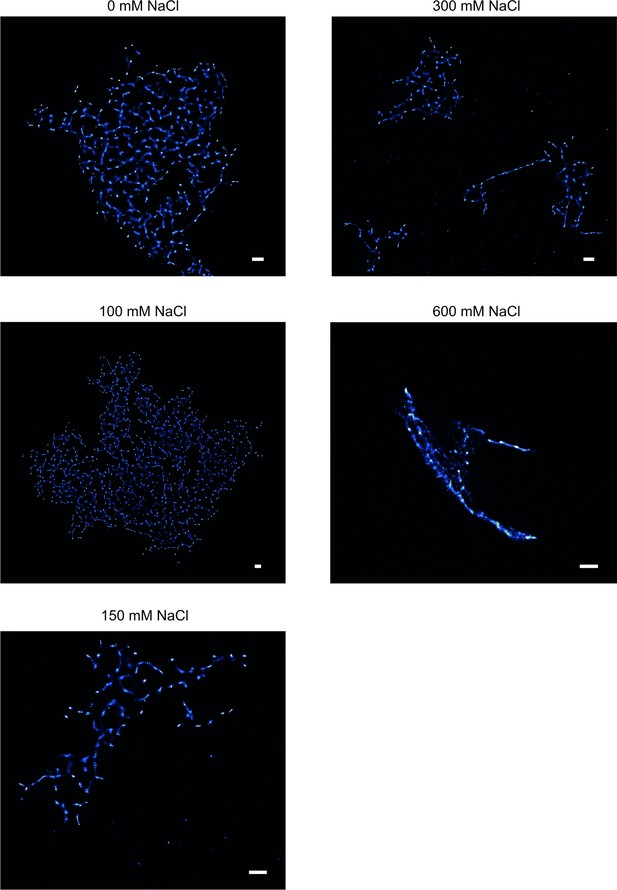
TAB SR microscopy images of aggregation endpoints of hPrP23 co-aggregated with syntaxin-6.
Native aggregation assay (NAA) was performed for hPrP23/syntaxin-6 co-aggregation (1:0.1 molar ratio) at increasing salt concentrations from 0 mM to 600 mM NaCl for 140 hr; scale bars 2 µm.
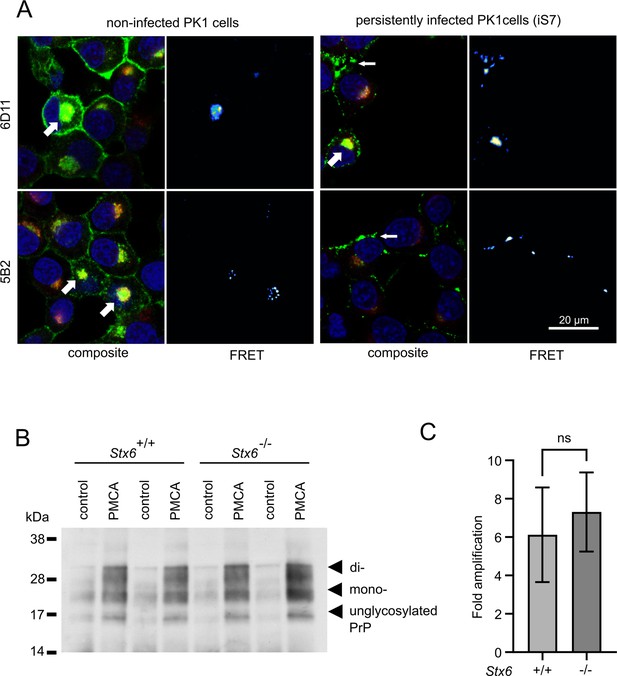
Interaction of PrP and syntaxin-6 in vivo.
(A) Non-infected PK1 cells and persistently infected PK1 cells (iS7) were immuno-stained with anti-PrP antibodies 6D11 and 5B2 (green), with anti-syntaxin-6 antibody (red) and with DAPI (blue). Förster resonance energy transfer (FRET) analysis reveals interaction in perinuclear compartments (wide arrows) and at membranes in infected cells (narrow arrows). Panels show zoomed regions of images in Figure 4—figure supplement 1. (B, C) In vitro prion replication by protein misfolding cyclic amplification (PMCA) using Stx6+/+ and Stx6-/- mouse brains as substrate. PMCA reactions were seeded with RML prions from terminally ill mice and subjected to PMCA for 96 cycles over 48 hr. (B) Representative western blot (ICSM35) after PK digestion. Molecular weight markers are indicated on the left. (C) The PrPSc signal was quantified using densitometry and normalized to the control unamplified reaction. Bar graphs each represent mean ± SD of biological replicates from three separate mice, each blotted as two technical replicates. Source data is available at https://doi.org/10.17632/yggpkrgnx8.1.
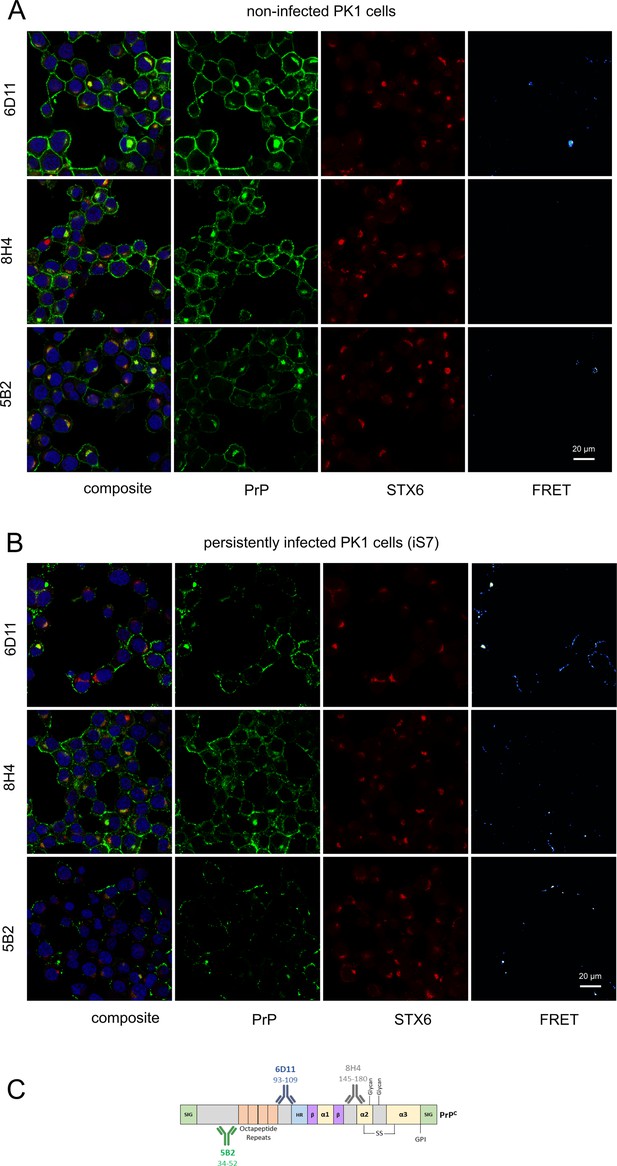
hPrP/syntaxin-6 Förster resonance energy transfer (FRET) analysis.
(A, B) PixFRET analysis of non-infected PK1 cells and persistently infected PK1 cells (iS7). Cells were immuno-stained with anti-PrP antibodies 6D11, 8H4, or 5B2 (green), with anti-syntaxin-6 antibody (STX6, red) and with DAPI. (C) Schematic of binding locations of antibodies used in FRET analysis (5B2, 6D11, 8H4). Numbers represent putative epitopes. (C) was adapted from Figure 2B from Castle and Gill, 2017.
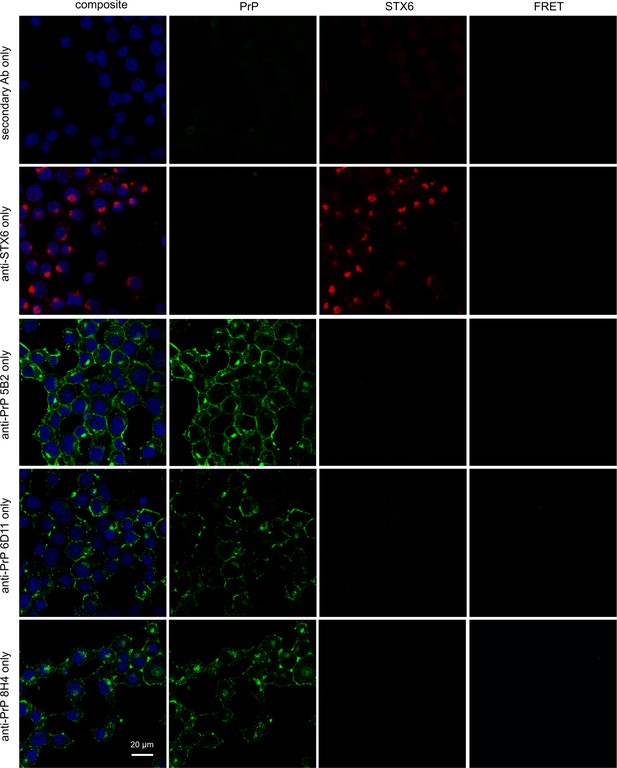
hPrP/syntaxin-6 Förster resonance energy transfer (FRET) analysis controls.
PixFRET analysis of control cells. iS7 cells were immuno-stained with anti-PrP antibodies 6D11, 8H4, or 5B2 (green) only, with anti-syntaxin-6 antibody (STX6, red) only, or with secondary antibodies only and with DAPI.
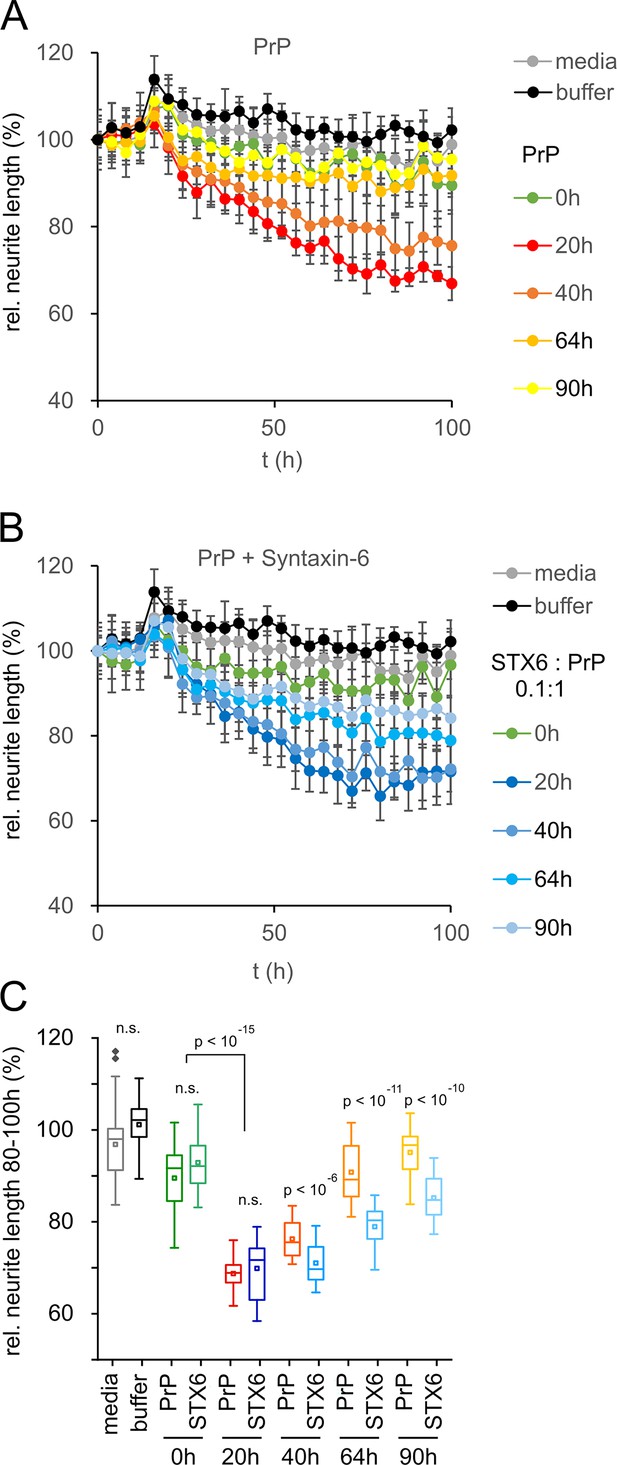
Toxicity in mouse primary neurons incubated with hPrP23/syntaxin-6 at different aggregation time points.
Native aggregation assay (NAA) was performed as in Figure 2 and samples were diluted 1:10 into cell culture media at the indicated time points (0 hr, 20 hr, 40 hr, 64 hr, 90 hr). (A) Plot of effect of PrP (250 nM) on neurite length compared to effect of media or NAA assay buffer at the same dilution from four independent wells. (B) Plot of effect of hPrP23 co-aggregated with syntaxin-6 (STX6, 1:10 molar ratio). (C) Box plot of average relative neurite lengths between 80–100 hr incubation of PrP in the cell culture media. p-Values were derived from ANOVA testing between PrP vs. PrP/syntaxin-6 samples at each time point.
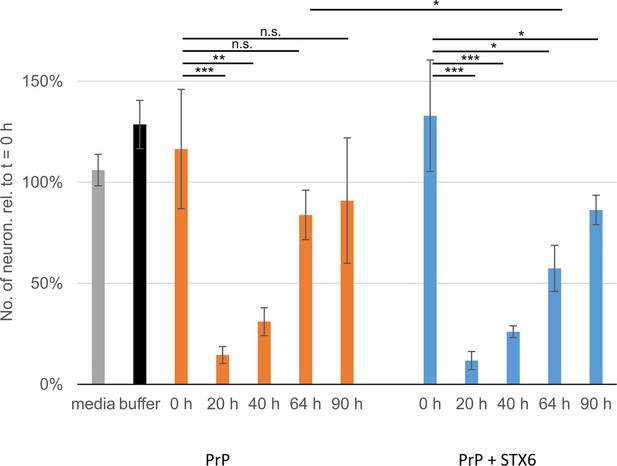
Survival of primary neurons.
Numbers of live primary neurons after incubation with hPrP23 or hPrP23 co-aggregated with syntaxin-6 (STX6, 1:10 molar ratio) as in Figure 5. Relative numbers of neurons after 4 hr incubation in four fields of view per sample were normalized to the same FOV at t = 0 hr; *p<0.05, **p<0.001, ***p<0.0001, n.s. not significant from ANOVA statistical analysis.
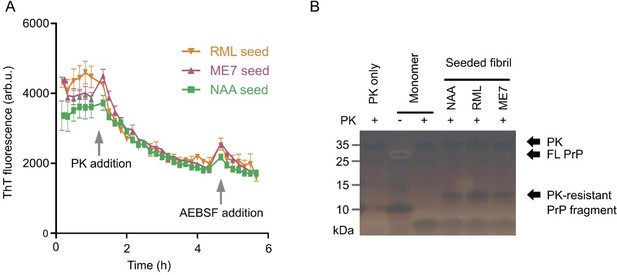
Murine (mPrP23) fibrils formed in NAA show limited proteinase K resistance.
(A) Baseline-corrected ThT fluorescence of endpoint NAA fibrils digested by PK (5 µg / mL). NAA fibrils had been formed in NAA assays seeded with recombinant fibrils (NAA), RML or ME7 prion rods. A ThT fluorescence baseline was recorded for 1.5 h, PK was added and AEBSF protease inhibitor was then added after 5 h incubation. Curves represent means ± SD of triplicate experiments. (B) Silver stained SDS-PAGE of the samples from panel A.
Additional files
-
Reporting standard 1
ARRIVE E10 checklist.
- https://cdn.elifesciences.org/articles/83320/elife-83320-repstand1-v2.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83320/elife-83320-mdarchecklist1-v2.pdf





