Plasmodium infection disrupts the T follicular helper cell response to heterologous immunization
Figures

DCs capture only a small fraction of infected RBCs in vivo.
Red blood cells infected with Plasmodium chabaudi (iRBCs) were enriched from infected mice, labeled with a fluorescent dye, and injected intravenously into naïve C57Bl/6 J mice. Labeled naïve RBCs (nRBCs) were injected as a control. Splenocytes were analyzed 30 min later for fluorescent iRBC label by flow cytometry. (A) Representative gating of bulk dendritic cells (DCs), B cells, and red pulp macrophages (RPMs). (B) Representative plots showing the distribution of labeled iRBCs in DCs, B cells, and RPMs. (C) Quantification showing the percentage of the total RBC+ population that consisted of DCs, RPMs, or B cells as indicated. (D) Quantification of RBC+ DCs, RPMs, and B cells expressed as a percentage of total splenocytes. A and B depict representative plots from one of four independent experiments. C and D show pooled data (mean +/- SD) from all four experiments, with each symbol representing one mouse (n=12 mice for iRBC treatment, 14 for nRBC treatment). **, p<0.01 and ****, p<0.0001 by one-way ANOVA with Tukey’s post-test. n.s., not significant.
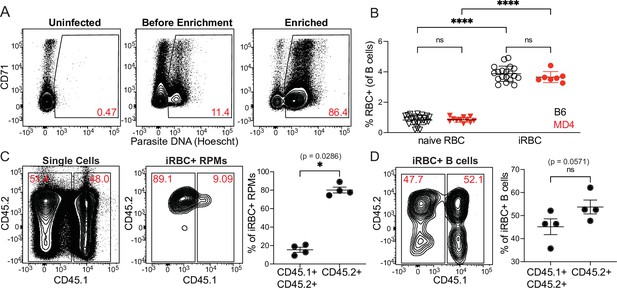
Uptake of enriched iRBCs by B cells.
(A) Infected RBCs were enriched over a magnetic column and degree of enrichment was assessed by flow cytometry. An uninfected sample is shown for comparison to pre-and post-enrichment samples from an infected mouse. Plots shown are pre-gated on Ter119+ CD45+ cells; then iRBCs are identified by staining of nucleic acids with Hoescht. CD71 is used to identify reticulocytes. (B) B6 or MD4 splenocytes were incubated with labeled naive RBCs or enriched iRBCs and RBC binding to B cells was assessed by flow cytometry. (C–D) To test whether splenocyte binding of iRBCs occurs in vivo or during tissue processing, labeled iRBCs were injected into CD45.2+ mice. Each spleen was harvested after 30min and disrupted along with a spleen from an untreated CD45.1+ CD45.2+ mouse. Binding of iRBCs to splenic CD45+ cells was assessed by flow cytometry. True in vivo uptake should be observed only in CD45.2+ cells, whereas binding that occurs during tissue processing should be distributed between CD45.2+ and CD45.1+ CD45.2+ splenocytes. (C) Contribution of each donor spleen to the total splenocyte pool (left panel) and subset of iRBC+ RPMs (middle panel). iRBC+ RPMs from each donor spleen are quantified (right panel). (D) Flow plot (left) and quantification (right) of the proportion of iRBC+ B cells derived from each donor spleen. A depicts representative plots from one of many enrichments. B-D show representative plots and pooled quantification from two independent experiments. *, p<0.05 and ****, p<0.0001 by one-way ANOVA with Tukey’s post-test (B) or Mann-Whitney (C, D). n.s., not significant.
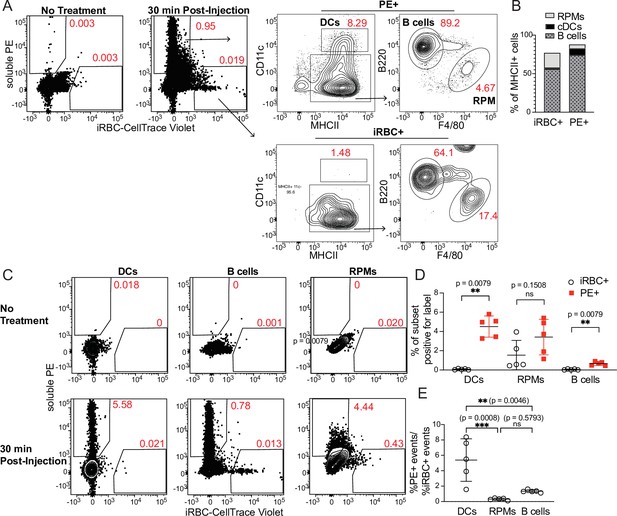
DCs take up soluble antigen more efficiently than infected RBCs.
Mice were intravenously injected with enriched, fluorescently labeled iRBCs together with PE and splenocytes were analyzed 30 min later for acquisition of fluorescent signal. (A) Representative gating of PE+ or iRBC+ splenocytes, further subsetted into the indicated APC populations. (B) Quantification of the distribution of PE and iRBC labeling among DCs, B cells, and RPMs. (C) Alternative gating strategy showing DCs, B cells, and RPMs in untreated mice (top row) and 30 min after injection of PE and iRBCs (bottom panel). (D) Quantification showing the percentage of each APC subset that acquired iRBC or PE label, as gated in (C). (E) Ratio of the percentage of each subset that acquired PE signal to the percentage of that subset that acquired iRBC signal. (B, D, and E) show pooled data (mean +/- SD) from two independent experiments (n=5 per group). **, p<0.01 and ***, p<0.001 by Mann-Whitney (D) or one-way ANOVA with Tukey’s post-test (E). n.s., not significant.
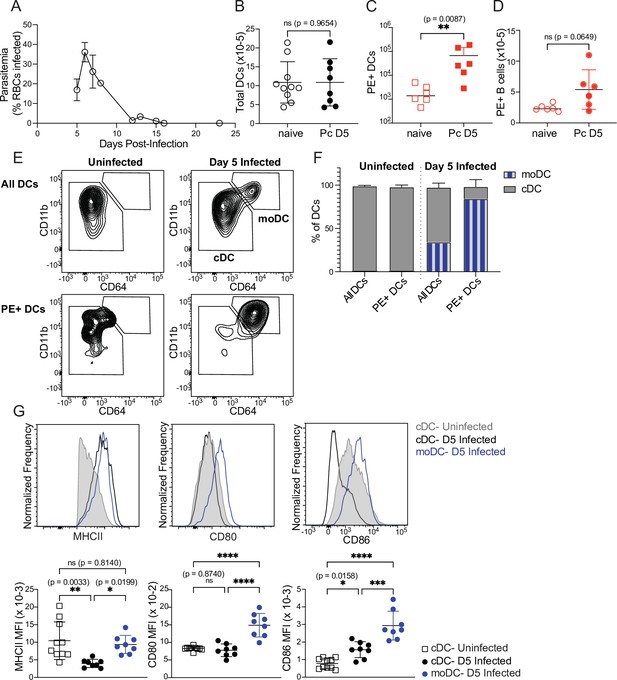
Monocyte-derived DCs take up majority of soluble antigen in Plasmodium-infected mice.
(A) Mice were infected with 106 P. chabaudi-parasitized RBCs and parasitemia was monitored by thin blood smear. (B) DCs were enumerated by flow cytometry in the spleens of mice at homeostasis or 5 days after P. chabaudi infection (Pc D5). (C) Naive or day 5-infected mice were injected intraperitoneally (i.p.) with PE, and PE+ splenic DCs were quantified after 30 min by flow cytometry. Note log scale. (D) Mice were treated as in C, and PE+ splenic B cells were quantified after 30 minutes. (E) Representative gating of conventional CD64- DCs (cDC) and CD11bhi CD64+ monocyte-derived DCs (moDC) in naïve or infected mice injected with PE. Top plots show all DCs; bottom plots show PE+ DCs. (F) Plots depicting the percentage of total or PE+ DCs that are cDC or moDC in naïve or infected mice. (G) Histograms and quantification showing expression (median fluorescent intensity, MFI) of MHCII, CD80, and CD86 on cDC and moDC. moDC are shown only from infected mice due to their scarcity in naïve mice. A shows data from one experiment (n=3), representative of many. B-D, F,and G show mean +/- SD from at least three experiments (n=6 per group for C, D; 10 uninfected, 8 infected for all others). *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001 by Mann-Whitney (B–D) or one-way ANOVA with Tukey’s post-test (G). n.s., not significant.
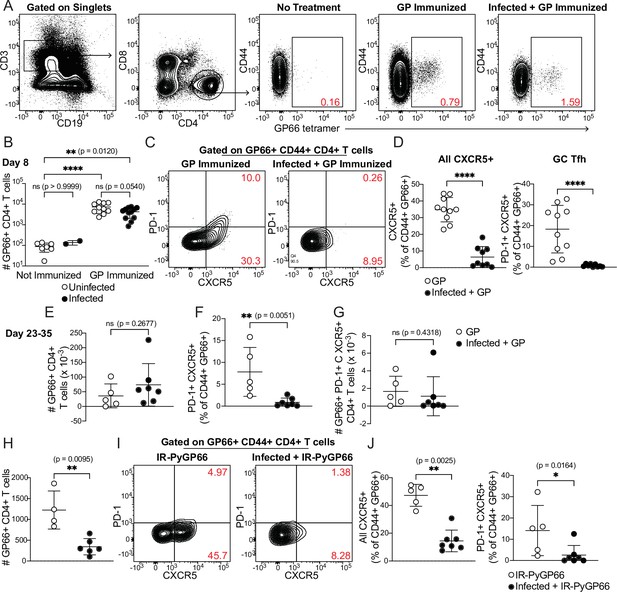
CD4+ T cell expansion, but disrupted polarization, following heterologous immunization in infected mice.
(A–G) Mice were immunized i.p. with recombinant LCMV glycoprotein (GP) plus adjuvant, either alone (GP Immunized) or 5 days after P. chabaudi infection (Infected +GP Immunized). GP66-specific CD4+ T cells were analyzed in spleen 8 days (A–D) or 23–35 days (E–G) after immunization. (A) Representative gating and (B) quantification of GP66+ CD4+ T cells from uninfected and infected mice with or without GP immunization. Note log scale in B. (C) Gating and (D) quantification of activated Tfh (CD44hi CXCR5+) and GC Tfh (CD44hi CXCR5+ PD-1hi) GP66-specific cells. (E–G) Total GP66-specific CD4+ T cells (E) and the frequency (F) and absolute number (G) of GP66-specific GC Tfh cells were quantified 23–35 days post-immunization. (H–J) Mice left uninfected or infected for 5 days with P. chabaudi were immunized with irradiated P. yoelii-parasitized RBCs expressing GP66 (IR-PyGP66), and splenic T cells were analyzed 8 days later. (H) Quantification of total GP66-specific CD4+ T cells. (I) Representative flow plot of CXCR5 and PD-1 expression on GP66-specific T cells. (J) Frequencies of GP66-specific Tfh cells (left graph) and GC Tfh cells (right graph). A-D represent data from four independent experiments (pooled n=10 “GP” mice, 12 “Infected +GP” mice). E–G show pooled results from one Day 23 experiment and one Day 35 experiment (n=5 “GP” mice, 7 “Infected +GP” mice). H–J show pooled results from two independent experiments (n=5 immunized, 7 infected +immunized). Red numbers within flow plot gates represent the frequency of cells within the gate. *, p<0.05, **, p<0.01 and ****, p<0.0001 by one-way ANOVA with Tukey’s post-test (B) or Mann-Whitney (others). n.s., not significant.

Abundant CXCR5 expression by bulk CD4+ T cells in infected mice.
Representative flow plot showing CXCR5 and PD-1 expression on bulk CD19- CD44hi CD4+ T cells in mice infected for 13 days with P. chabaudi. This timepoint corresponds to the time of analysis for GP66-specific T cells in mice immunized with GP 5 days after P. chabaudi infection, then harvested 8 days after immunization (Figure 4).
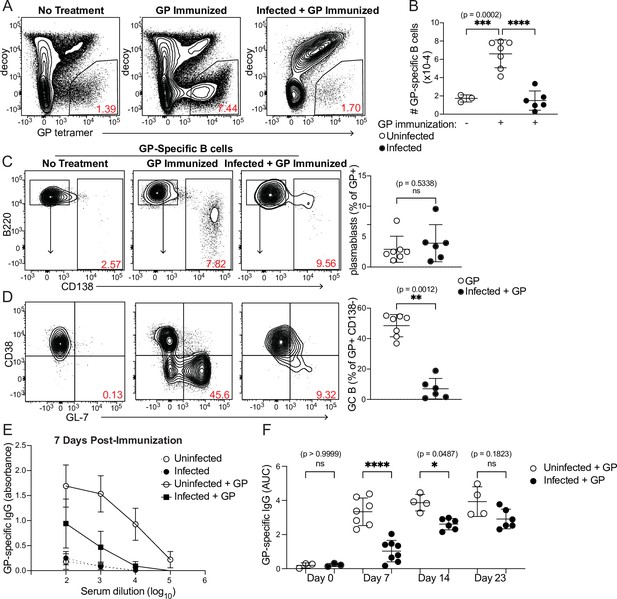
Curtailed B cell expansion and antibody production in infected mice following heterologous immunization.
Mice were immunized with or without concurrent P. chabaudi infection, as in Figure 4. (A) Representative gating and (B) quantification of GP-specific splenic B cells 8 days post-immunization. In A, plots were pre-gated on B cells. (C, D) Gating (left panels) and quantification (right) of CD138+ plasmablasts (C) or CD38- GL7+ GC B cells (D) 8 days after immunization with GP. Plots in D were pre-gated on CD138- cells. (E) GP-specific IgG antibody was measured by ELISA in the serum of uninfected or infected mice without immunization or 7 days after immunization with GP. (F) GP-specific serum antibodies (expressed as Area Under the Curve, AUC) measured by ELISA at the indicated times post-immunization. A-D represent data from three independent experiments (n=7 GP, 6 Infected + GP); E and F are pooled from two experiments. Red numbers within flow plot gates represent the frequency of cells within the gate. *, p< 0.05, **, p < 0.01, ***, p < 0.001, and ****, p < 0.0001 by one-way ANOVA with Tukey’s post-test (B, F) or Mann-Whitney (others). n.s., not significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| antibody | Anti-B220 (RA3-6B2), rat monoclonal | BD | Cat# 563892; RRID:AB_2738470 | 1:100 dilution |
| antibody | Anti-CD138 (281-2), rat monoclonal | Biolegend | Cat# 142516; RRID:AB_2562337 | 1:100 dilution |
| antibody | Anti-CD38 (90), rat monoclonal | Biolegend | Cat# 102717; RRID:AB_2072892 | 1:100 dilution |
| antibody | Anti-GL7 (GL-7), rat monoclonal | eBioscience | Cat# 48-5902-80; RRID:AB_10854881 | 1:100 dilution |
| antibody | Anti-IgM (RMM-1), rat monoclonal | Biolegend | Cat# 406512; RRID:AB_2075943 | 1:100 dilution |
| antibody | Anti-CD4 (GK1.5), rat monoclonal | eBioscience | Cat# 19-0041-82; RRID:AB_469533 | 1:100 dilution |
| antibody | Anti-CD8 (53–6.7), rat monoclonal | BD | Cat# 553034; RRID:AB_394572 | 1:100 dilution |
| antibody | Anti-CD11b (M1/70), rat monoclonal | eBioscience | Cat# 14-0112-81; RRID:AB_467107 | 1:100 dilution |
| antibody | Anti-CD86 (GL-1), rat monoclonal | Biolegend | Cat# 105013; RRID:AB_439782 | 1:100 dilution |
| antibody | Anti-CD45.1 (A20), mouse monoclonal | BD | Cat# 560578; RRID:AB_1727488 | 1:100 dilution |
| antibody | Anti-CD45.2 (104), mouse monoclonal | BD | Cat# 560696; RRID:AB_1727494 | 1:100 dilution |
| antibody | Anti-CD11c (HL3), Armenian hamster monoclonal | BD | Cat# 553802; RRID:AB_395061 | 1:100 dilution |
| antibody | Anti-CD16/CD32 (2.4G2), rat monoclonal | BD | Cat# 553142 RRID:AB_394657 | 1:1000 dilution |
| antibody | Anti-CD64 (X54-5/7.1), mouse monoclonal | Biolegend | Cat# 139309 RRID:AB_2562694 | 1:100 dilution |
| antibody | Anti-Thy1.2 (53–2.10, rat monoclonal) | BD | Cat# 565257 RRID:AB_2739136 | 1:100 dilution |
| antibody | Anti-MHCII (M5/114.15.2), rat monoclonal | eBioscience | Cat# 56-5321-82 RRID:AB_494009 | 1:100 dilution |
| antibody | Anti-F4/80 (BM8), rat monoclonal | Biolegend | Cat# 123115 RRID:AB_893493 | 1:100 dilution |
| antibody | Anti-Siglec H (551), rat monoclonal | Biolegend | Cat# 129611 RRID:AB_10643574 | 1:100 dilution |
| antibody | Anti-CD3e (500A2), Syrian hamster monoclonal | BD | Cat# 553240 RRID:AB_394729 | 1:100 dilution |
| antibody | Anti-CD80 (16–10 A1), Armenian hamster monoclonal | eBioscience | Cat# 12-0801-82 RRID:AB_465752 | 1:100 dilution |
| antibody | Anti-CD19 (1D3), rat monoclonal | eBioscience | Cat# 47-0193-82 RRID:AB_10853189 | 1:100 dilution |
| antibody | Anti-CD44 (IM7), rat monoclonal | BD | Cat# 560567 RRID:AB_1727480 | 1:100 dilution |
| antibody | Anti-CD62L (MEL-14), rat monoclonal | Biolegend | Cat# 104440 RRID:AB_2629685 | 1:100 dilution |
| antibody | Anti-PD-1 (J43), Armenian hamster monoclonal | eBioscience | Cat# 11-9985-85 RRID:AB_465473 | 1:100 dilution |
| antibody | Anti-CXCR5 (2G8), rat monoclonal | BD | Cat# 560617 RRID:AB_1727521 | 1:20 dilution |
| antibody | Anti-CD71 (R17217), rat monoclonal | Invitrogen | Cat# 12-0711-83 RRID:AB_465741 | 1:200 dilution |
| antibody | Anti-CD45 (30-F11), rat monoclonal | BD | Cat# 559864 RRID:AB_398672 | 1:200 dilution |
| antibody | Anti-Ter119 (Ter119), rat monoclonal | Invitrogen | Cat# MA5-17822 RRID:AB_2539206 | 1:200 dilution |
| Chemical compound, drug | Hoescht 33342 Solution | ThermoFisher | Cat# 62249 | 1:1000 dilution |
| Peptide, recombinant protein | LCMV Glycoprotein | E Ollman Saphire | N/A | |
| Peptide, recombinant protein | I-A(b) LCMV GP 66–77 Monomer | NIH Tetramer Core Facility | N/A | Used at 1 uM |
| Peptide, recombinant protein | Streptavidin-APC | Agilent | Cat# PJ27S-1 | |
| Strain, strain background (Plasmodium yoelii) | Plasmodium yoelii (17 X NL) GP66 | Hahn et al., 2018 | N/A | |
| Strain, strain background (Plasmodium chabaudi) | Plasmodium chabaudi AS | MR4 Stock Center | #MRA-743 | |
| Strain, strain background (Mus musculus) | C56Bl/6 J (B6) mice | Jackson Labs | Stock #000664; RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mus musculus) | CD45.1+mice | Jackson Labs | Stock #002014; RRID:IMSR_JAX:002014 | |
| Strain, strain background (Mus musculus) | C57BL/6-Tg(IghelMD4)4Ccg/J (MD4) mice | Jackson Labs | Stock #002595; RRID:IMSR_JAX:002595 | |
| Software, algorithm | Prism 9 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 | |
| Software, algorithm | FlowJo 10 | TreeStar | RRID:SCR_008520 |





