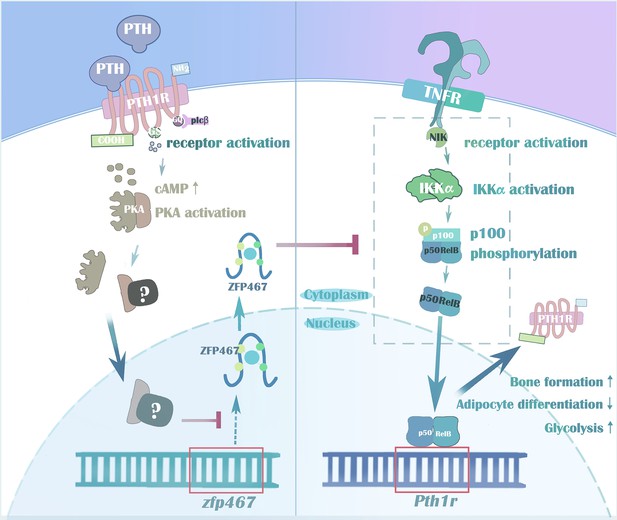
PTH regulates osteogenesis and suppresses adipogenesis through Zfp467 in a feed-forward, PTH1R-cyclic AMP-dependent manner
Figures
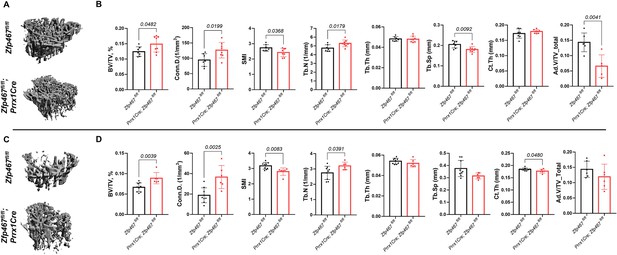
Prrx1Cre Zfp467 mice have more trabecular bone mass and less adipose tissue in bone marrow, recapitulating the global Zfp467 null mice.
(A, B) Male and (C, D) female 12-week-old Prrx1Cre; Zfp467fl/fl mice and control mice were measured using trabecular and cortical bone of tibiae. Marrow adipose tissue volume (Ad.V) was quantified by osmium tetroxide staining and micro-computed tomography (μCT). Data shown as mean ± SD by unpaired Student’s t test, n=5–8 per group.
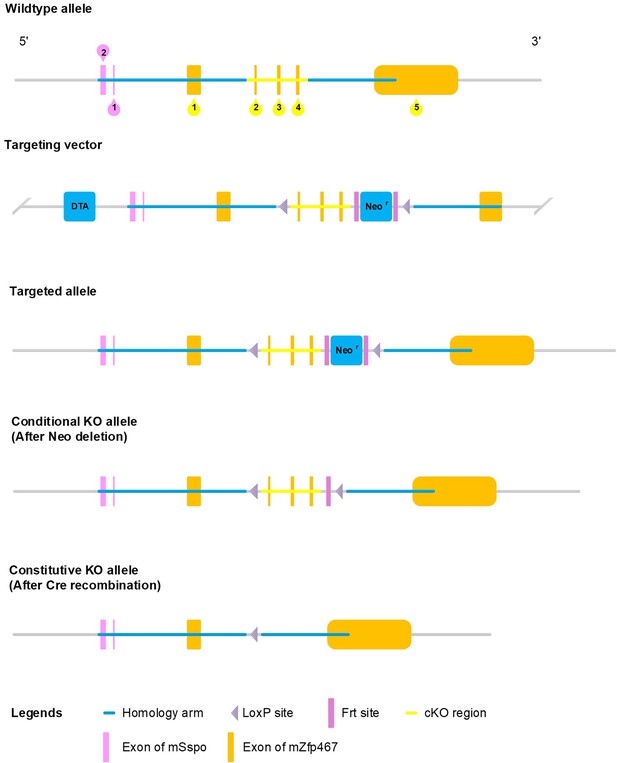
Generation of Zfp467 flox mice.
Exons 2–4 was selected as conditional knockout region. The targeting vector was generated by PCR using BAC clone RP24-144J8 and RP23-24K23 from the C57BL/6J library as template.
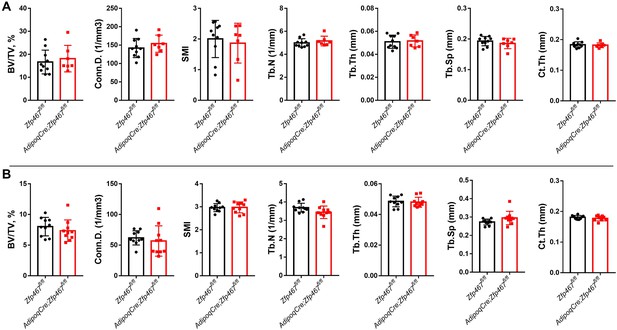
AdipoqCre Zfp467 have similar trabecular and cortical bone mass with controls.
(A) Male and (B) female 12-week-old mice and control Zfp467fl/fl; AdipoqCre mice were measured using trabecular and cortical bone of tibiae. Data shown as mean ± SD by unpaired Student’s t test, n=7–10 per group.
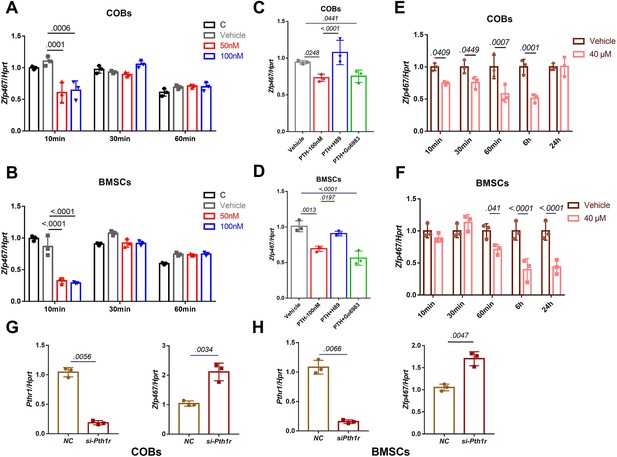
Parathyroid hormone (PTH) suppressed the expression levels of Zfp467 via the PKA pathway.
(A) PTH treatments significantly suppressed Zfp467 expression within 10 min of treatment in Zfp467+/+ calvarial osteoblasts (COBs). Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. (B) PTH treatments significantly suppressed Zfp467 expression within 10 min of treatment in Zfp467+/+ bone marrow stromal cells (BMSCs). Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. (C) qPCR results· of Zfp467+/+ in COBs with 2 hr PKA or PKC inhibitor treatment prior to 10 min of 100 nM PTH exposure, PKA but not PKC inhibitor was able to rescue the suppression of Zfp467 induced by PTH. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. (D) qPCR results of Zfp467+/+ in BMSCs with 2 hr PKA or PKC inhibitor treatment prior to 10 min of 100 nM PTH exposure. PKA but not PKC inhibitor was able to rescue the suppression of Zfp467 induced by PTH. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. Forskolin significantly suppressed Zfp467 expression within 1 hr of treatment in Zfp467+/+ COBs. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (F) Forskolin significantly suppressed Zfp467 expression after 6 hr of treatment in Zfp467+/+ BMSCs. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (G) Pth1r-siRNA treatment in Zfp467+/+ COBs led to an increase of Zfp467 expression in Zfp467+/+ COBs. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (H) Pth1r-siRNA treatment in Zfp467+/+ COBs led to an increase of Zfp467 expression in Zfp467+/+ BMSCs. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. NC, negative control.
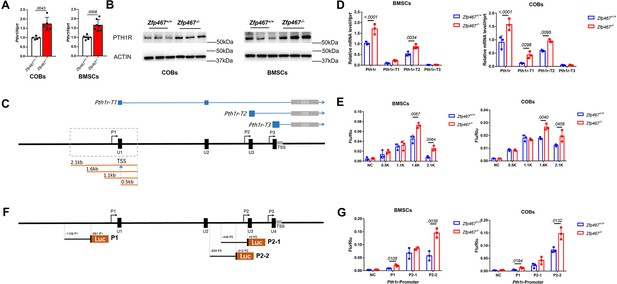
Zfp467-/- cells have greater Pth1r transcriptional levels driven by both the P1 and P2 promoter.
(A) qPCR results of baseline calvarial osteoblasts (COBs) and bone marrow stromal cells (BMSCs). Higher expression level of Pth1r was found in both Zfp467-/- COBs and BMSCs. Data shown as mean ± SD by unpaired Student’s t test, n=5–7 independent experiments for each group. (B) Western blot analysis of baseline COBs and BMSCs. Higher expression level of PTH1R was found in both Zfp467-/- COBs and BMSCs. (C) A schematic of three different Pth1r transcripts and P1 promoter of Pth1r. Four different length P1 promoter constructs were designed and inserted into dual-fluorescence reporter vector. (D) qPCR results of three Pth1r transcripts and total Pth1r. Total Pth1r and Pth1r-T1, T2 but not Pth1r-T3 were upregulated in both Zfp467-/- COBs and BMSCs. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (E) Dual-fluorescence assay using indicated four P1 reporter constructs. The 1.6 and 2.1 kb constructs-driven reporter is higher activated in Zfp467-/- cells compared to Zfp467+/+ cells. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (F) A schematic of P1 and P2 promoter constructs of Pth1r. (G) Dual-fluorescence assay using indicated P1 and P2 reporter constructs. Both P1 and P2-2 were found significantly higher activated in Zfp467-/- cells. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. NC, negative control, TSS, transcription starting site.
-
Figure 3—source data 1
Western blot for Figure 3B.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig3-data1-v2.zip
-
Figure 3—source data 2
Western blot for Figure 3B PTH1R in calvarial osteoblasts (COBs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig3-data2-v2.zip
-
Figure 3—source data 3
Western blot for Figure 3B ACTIN in calvarial osteoblasts (COBs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig3-data3-v2.zip
-
Figure 3—source data 4
Western blot for Figure 3B PTH1R in bone marrow stromal cells (BMSCs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig3-data4-v2.zip
-
Figure 3—source data 5
Western blot for Figure 3B ACTIN in bone marrow stromal cells (BMSCs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig3-data5-v2.zip
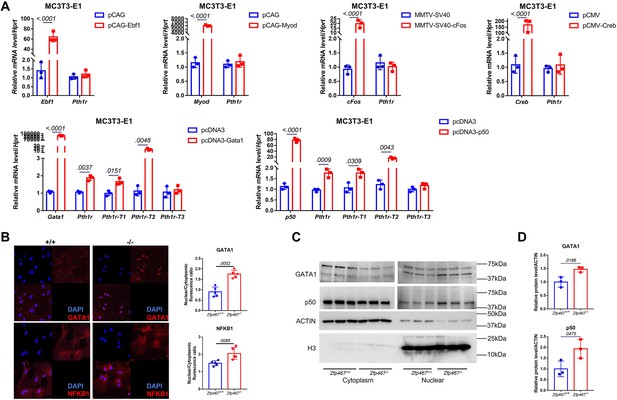
Zfp467-/- cells have higher NFκB1 and GATA1 nuclear translocation.
(A) qPCR results of overexpression of Ebf1, Myod, Myog, Gata1, and NFκB1 in MC3T3-E1 cell line. GATA1 and NFκB1 overexpression could significantly upregulate the expression level of Pth1r. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (B) Representative confocal images of GATA1 and NFκB1 immunofluorescence in Zfp467+/+ and Zfp467-/- bone marrow stromal cells (BMSCs) and related quantification. (C) Nuclear protein level of GATA1 and NFκB1 in Zfp467+/+ and Zfp467-/- BMSCs. (D) Quantification analysis for nuclear protein level of GATA1 and NFκB1 in Zfp467+/+ and Zfp467-/- BMSCs. Data shown as mean ± SD, n=3 independent experiments for each group. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group.
-
Figure 4—source data 1
Western blot for Figure 3C.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data1-v2.zip
-
Figure 4—source data 2
Western blot for Figure 3C ACTIN.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data2-v2.zip
-
Figure 4—source data 3
Western blot for Figure 3C H3.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data3-v2.zip
-
Figure 4—source data 4
Western blot for Figure 3C p50 in cytoplasm.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data4-v2.zip
-
Figure 4—source data 5
Western blot for Figure 3C GATA1 in cytoplasm.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data5-v2.zip
-
Figure 4—source data 6
Western blot for Figure 3C GATA1 in nuclear.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data6-v2.zip
-
Figure 4—source data 7
Western blot for Figure 3C p50 in nuclear.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig4-data7-v2.zip
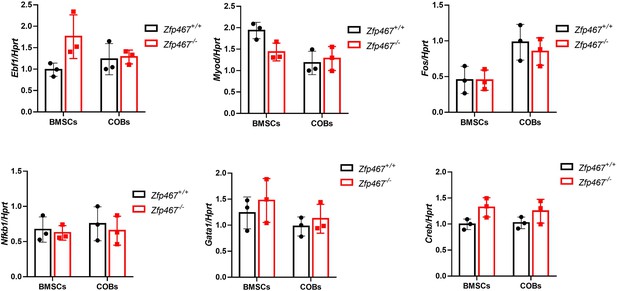
qPCR results of expression of Ebf1, Myod, Fos, Gata1, Nfkb1, and Creb in Zfp467+/+ and Zfp467-/- calvarial osteoblasts (COBs) and bone marrow stromal cells (BMSCs).
Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group.
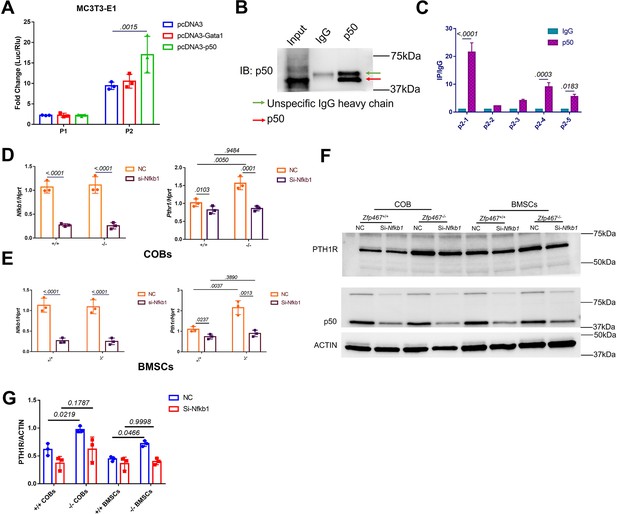
NFκB1 was found to transactivate Pth1r expression in Zfp467-/- cells.
(A) Reporter assays using the indicated P1 or P2 reporter construct and an expression vector bearing Gata1, Nfkb1, or a control empty vector. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. (B) Immunoblot assay using a control rabbit IgG antibody (IgG) or the anti-NFκB1 antibody during chromatin immunoprecipitation assay. (C) DNA enrichment of Pth1r P2 promoter, ratio between NFκB1 and IgG IP products, first part and last two parts of P2 were significantly enriched by NFκB1 antibody. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (D, E) qPCR results of the expression levels of Nfkb1 and Pth1r in Nfkb1 siRNA-treated Zfp46 +/+ and Zfp467-/- calvarial osteoblasts (COBs) and bone marrow stromal cells (BMSCs). Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. (F) Western blot analysis of Nfkb1 and Pth1r in Nfkb1 siRNA-treated Zfp467 +/+ and Zfp467-/- COBs and BMSCs. (G) Quantification for PTH1R protein level. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. NC, negative control.
-
Figure 5—source data 1
Western blot for Figure 5B and F.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-data1-v2.zip
-
Figure 5—source data 2
Western blot for Figure 5B p50 with IP samples.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-data2-v2.zip
-
Figure 5—source data 3
Western blot for Figure 5C PTH1R.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-data3-v2.zip
-
Figure 5—source data 4
Western blot for Figure 5C p50.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-data4-v2.zip
-
Figure 5—source data 5
Western blot for Figure 5C ACTIN.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-data5-v2.zip
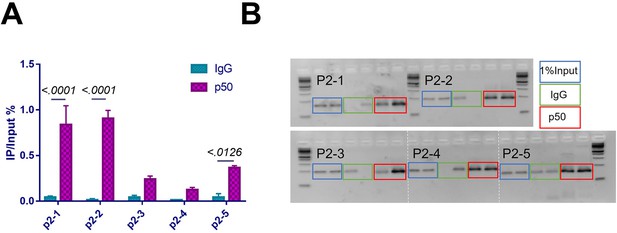
p2 promoter of Pth1r was properly enriched in p50-ChIP-qPCR assay using MC3T3-E1 nuclear extracts.
(A) DNA enrichment of Pth1r P2 promoter, ratio between IP and input, first two parts of P2 were enriched using NFκB1 antibody. Data shown as mean ± SD by unpaired Student’s t test, n=3 replicates for each group. (B) RT-PCR product of chromatin immunoprecipitation assay using the nuclear extracts from MC3T3-E1 cells.
-
Figure 5—figure supplement 1—source data 1
Nucleic acid blot for Figure 5—figure supplement 1B.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-figsupp1-data1-v2.zip
-
Figure 5—figure supplement 1—source data 2
Nucleic acid blot for Figure 5—figure supplement 1B, PCR blot for Pth1r amplification product.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig5-figsupp1-data2-v2.zip
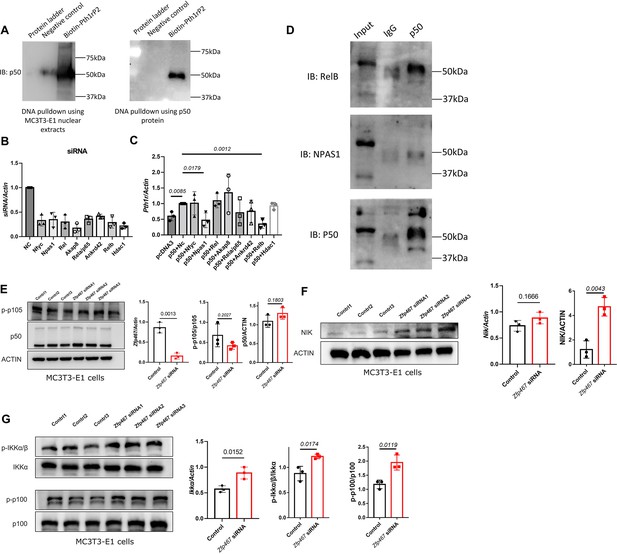
NFκB1 heterodimerize with RelB to transactivate the expression of Pth1r.
(A) DNA pulldown assay with biotin-labeled Pth1r P2. MC3T3-E1 nuclear extracts or NFκB1 recombinant protein was probed with biotin-Pth1rP2 and then subjected to immunoblotting using NFκB1 antibody. (B) qPCR results of the expression levels of Nfyc, Npas1, Rel, Akap8, Rela, Ankrd42, Relb, and Hdac1 in related siRNA-treated MC3T3-E1 cells. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group. NC, negative control. (C) qPCR results of the expression levels of Pth1r in Nfkb1 overexpression plasmid and Nfyc, Npas1, Rel, Akap8, Rela, Ankrd42, Relb, or Hdac1 siRNA co-transfected MC3T3-E1 cells. NC, negative control. (D) IP results using NFκB1 antibody in MC3T3-E1 protein extracts, IgG was used as a negative control. (E) Protein level of p-p105 and p50 in Zfp467 knockdown MC3T3-E1 cells. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (F) Protein and mRNA level of NFκB-inducing kinase (NIK) in control and Zfp467 siRNA-treated MC3T3-E1 cells. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (G) Protein level of p-IKKa and p-p100 and mRNA level of Ikka in control and Zfp467 siRNA-treated MC3T3-E1 cells. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group.
-
Figure 6—source data 1
Western blot for Figure 6A, D, E, F and G.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data1-v2.zip
-
Figure 6—source data 2
Western blot for Figure 6A p50 in DNA pulldown experiment using nuclear extract.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data2-v2.zip
-
Figure 6—source data 3
Western blot for Figure 6A p50 in DNA pulldown experiment using p50 purified protein.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data3-v2.zip
-
Figure 6—source data 4
Western blot for Figure 6D RelB.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data4-v2.zip
-
Figure 6—source data 5
Western blot for Figure 6D NPAS1.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data5-v2.zip
-
Figure 6—source data 6
Western blot for Figure 6D p50.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data6-v2.zip
-
Figure 6—source data 7
Western blot for Figure 6E p-p105.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data7-v2.zip
-
Figure 6—source data 8
Western blot for Figure 6E p50/p105.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data8-v2.zip
-
Figure 6—source data 9
Western blot for Figure 6E ACTIN.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data9-v2.zip
-
Figure 6—source data 10
Western blot for Figure 6F NFκB-inducing kinase (NIK).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data10-v2.zip
-
Figure 6—source data 11
Western blot for Figure 6F ACTIN.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data11-v2.zip
-
Figure 6—source data 12
Western blot for Figure 6F p-IKKα/β.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data12-v2.zip
-
Figure 6—source data 13
Western blot for Figure 6F IKKα.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data13-v2.zip
-
Figure 6—source data 14
Western blot for Figure 6G p-p100.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data14-v2.zip
-
Figure 6—source data 15
Western blot for Figure 6G p100.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig6-data15-v2.zip
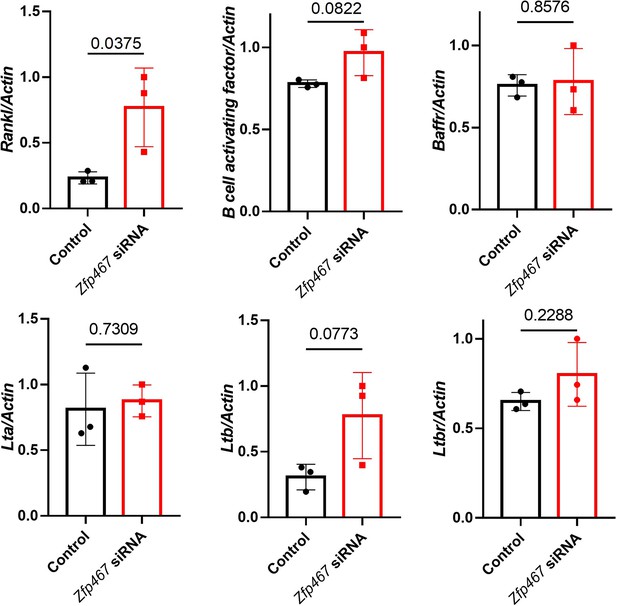
mRNA level of NFκB non-canonical pathway stimulus and related receptors in control and Zfp467 siRNA-treated MC3T3-E1 cells.
Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group.
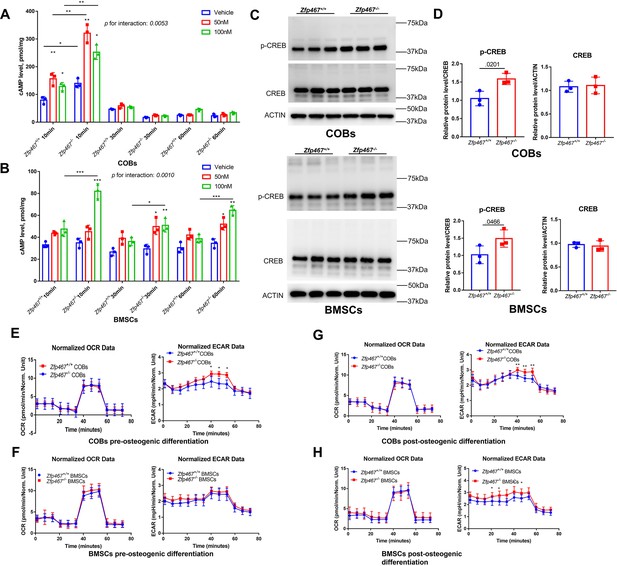
Zfp467-/- cells have increased parathyroid hormone (PTH) signaling and higher extracellular acidification rates.
(A) Cyclin adenosine monophosphate (cAMP) ELISA of undifferentiated calvarial osteoblasts (COBs), from 10 to 60 min with 100 nM PTH treatment, PTH increased cAMP expression within 10 min of treatment, and Zfp467-/- cells have higher level of cAMP than Zfp467+/+. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group; *, p<0.01, **, p<0.001. (B) cAMP ELISA of undifferentiated bone marrow stromal cells (BMSCs), from 10 to 60 min with 100 nM PTH treatment, Zfp467-/- BMSCs had significantly higher level of intracellular cAMP after 10–60 min exposure of PTH. Data shown as mean ± SD by one-way ANOVA, n=3 independent experiments for each group; *, p<0.01, **, p<0.001. (C, D) Western blot and quantitative analysis of pre-differentiated COBs and BMSCs. Higher expression levels of p-CREB but not total CREB was found in both Zfp467-/- COBs and BMSC. Data shown as mean ± SD by unpaired Student’s t test, n=3 independent experiments for each group. (E, F) Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) of undifferentiated Zfp467+/+ and Zfp467-/- COBs or BMSCs. No difference was found regarding OCR between genotypes, but Zfp467-/- BMSCs had higher ECAR than Zfp467+/+ BMSCs. Data shown as mean ± SD by unpaired Student’s t test, n=12 technical replicates. *, p<0.01, **, p<0.001. (G, H) OCR and ECAR of Zfp467+/+ and Zfp467-/- COBs or BMSCs after 3 days’ osteogenic differentiation. No difference was found regarding OCR between genotypes, but both Zfp467-/- COBs and BMSCs cells had significantly higher ECAR level than Zfp467+/+ cells. Data shown as mean ± SD by unpaired Student’s t test, n=12 technical replicates. *, p<0.01, **, p<0.001.
-
Figure 7—source data 1
Western blot for Figure 7C.
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data1-v2.zip
-
Figure 7—source data 2
Western blot for Figure 7C p-CREB in calvarial osteoblasts (COBs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data2-v2.zip
-
Figure 7—source data 3
Western blot for Figure 7C CREB in calvarial osteoblasts (COBs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data3-v2.zip
-
Figure 7—source data 4
Western blot for Figure 7C ACTIN in calvarial osteoblasts (COBs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data4-v2.zip
-
Figure 7—source data 5
Western blot for Figure 7C p-CREB in bone marrow stromal cells (BMSCs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data5-v2.zip
-
Figure 7—source data 6
Western blot for Figure 7C CREB in bone marrow stromal cells (BMSCs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data6-v2.zip
-
Figure 7—source data 7
Western blot for Figure 7C ACTIN in bone marrow stromal cells (BMSCs).
- https://cdn.elifesciences.org/articles/83345/elife-83345-fig7-data7-v2.zip
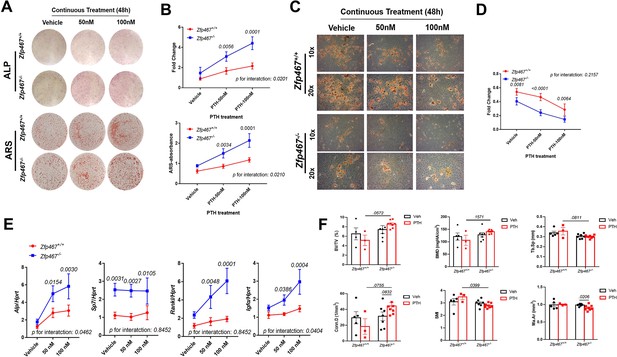
Zfp467-/-cells showed increased sensitivity to parathyroid hormone (PTH) and enhanced pro-osteogenic as well as anti-adipogenic effects.
(A) Representative images of alkaline phosphatase (ALP) (day 7) and alizarin red staining (ARS) (day 14) of differentiated calvarial osteoblasts (COBs) with PTH treatment. An increase in PTH dose led to an increase in ALP staining and mineralization in both Zfp467+/+ and Zfp467-/- COBs. Zfp467-/- COBs showed more ALP-positive cells and mineralization than Zfp467+/+ COBs. (B) ALP stain and ARS quantification in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. (C) Representative images of Oil Red O (ORO) staining (10× and 20×) of COBs after 14 days in’ osteogenic differentiation with PTH treatment. While PTH treatment inhibited adipocyte formation in Zfp467+/+ and Zfp467-/- groups, the Zfp467-/- group showed fewer adipocytes in all treatment groups as compared to Zfp467+/+. (D) ORO stain quantification in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. (E) qPCR results for osteoblast-related genes after 7 days’ osteogenic differentiation in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. (F) PTH treatment for female 12-week-old global Zfp467-/- mice mice and control mice were measured using trabecular and cortical bone of tibiae after 1 week PTH treatment. Data shown as mean ± SD by two-way ANOVA, n=5–8 per group.
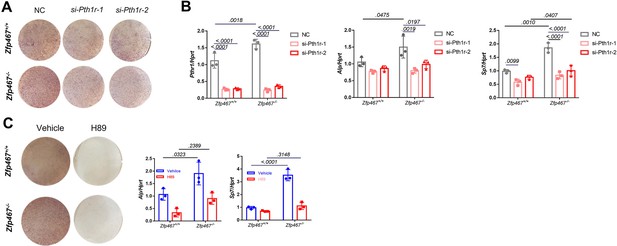
Gene silencing of Pth1r or PKA inhibitors suppressed Zfp467-/- induced increased osteogenic differentiation.
(A) Representative images of alkaline phosphatase (ALP) staining of differentiated calvarial osteoblasts (COBs) with Pth1r or control siRNA treatment. (B) qPCR results for osteogenic differentiation-related genes after 7 days’ osteogenic differentiation and siRNA treatment in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. (C) Representative images of ALP staining and qPCR results for osteogenic differentiation-related genes after 7 days’ osteogenic differentiation and PKA inhibitor treatment in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group. NC, negative control.
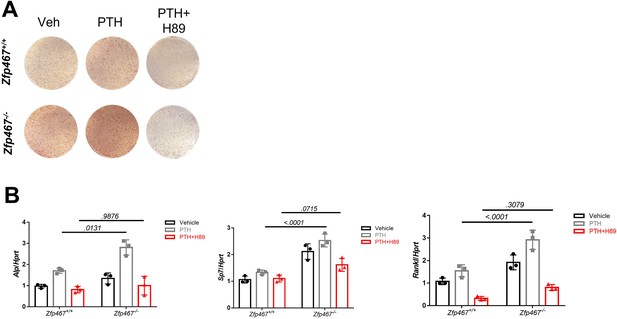
PKA inhibitor reversed the enhanced action of PTH on the osteogenic differentiation seen in Zfp467-/- COBs.
(A) Representative images of alkaline phosphatase (ALP) staining of differentiated calvarial osteoblasts (COBs) with 100 nM parathyroid hormone (PTH) treatment and PKA inhibitor. (B) qPCR results for osteogenic differentiation-related genes after 7 days’ osteogenic differentiation, PTH treatment and PKA inhibitor treatment in COBs. Data shown as mean ± SD by two-way ANOVA, n=3 independent experiments for each group.
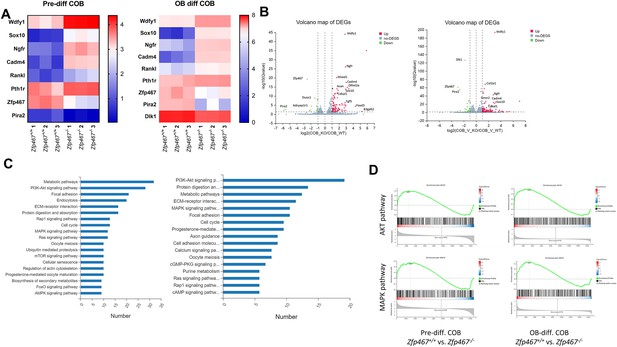
Wdfy1, Sox10, and Ngfr were found upregulated and MAPK, AKT pathways were found activated in Zfp467-/- calvarial osteoblasts (COBs).
(A) Heat map of differentially expressed genes (DEG) from COBs (Zfp467+/+ and Zfp467-/-) at pre-differentiation (Panel A) and at differentiation with a p-value < 0.05 and a fold change >2.0 or <–2.0. (B) These DEGs that represented by volcano plots. (C) Functional annotation (Cellular Component [CC]) for the DEGs for Zfp467+/+ vs Zfp467-/- for pre- (left) and post-differentiated COBs (right). (D) GSEA enrichment plots for AKT and MAPK pathways. (E) qRT-PCR was performed on COBs from Zfp467+/+ and -/- to confirm gene expression changes noted by RNA-seq. (F) Overexpression of Zfp467 in MC3T3-E1 cells confirmed statistically significant suppression of the top three genes (Wdfy1, Sox10, and Ngfr), when compared to GFP overexpression; p<0.05 or lower. Data shown as mean ± SD by unpaired Student’s t test, n=5–8 independent experiments for each group.
Tables
Quantification of structural and cellular parameters in the left tibiae of 12-week-old Prrx1Cre;Zfp467fl/fl and control Zfp467fl/fl mice by histomorphometry.
| Male | Female | |||||
|---|---|---|---|---|---|---|
| Zfp467fl/fl | Prrx1Cre; Zfp467fl/fl | p-Value | Zfp467fl/fl | Prrx1Cre; Zfp467fl/fl | p-Value | |
| BV/TV (%) | 9.1482±2.6523 | 12.570±2.9694 | 0.06 | 7.4561±1.9473 | 8.1315±3.8504 | 0.71 |
| Tb.Th (μm) | 35.961±3.7772 | 37.700±5.8760 | 0.63 | 35.384±2.5611 | 34.748±6.5862 | 0.76 |
| Tb.Sp (μm) | 396.80±168.61 | 274.83±62.817 | 0.13 | 484.18±119.44 | 501.35±288.61 | 0.90 |
| Tb.N (n/μm) | 2.5294±0.6037 | 3.3260±0.4942 | 0.03 | 2.0813±0.4175 | 2.3440±1.1650 | 0.61 |
| OS/BS (%) | 21.767±11.722 | 31.987±12.704 | 0.16 | 14.244±5.1159 | 8.8012±4.2810 | 0.07 |
| O.Th (μm) | 2.0084±0.4411 | 4.8227±3.4306 | 0.07 | 3.1650±0.4587 | 3.4638±1.2793 | 0.60 |
| Ob.S/BS (%) | 16.672±7.6973 | 25.817±8.5871 | 0.07 | 21.776±6.8886 | 18.210±8.0428 | 0.43 |
| N.Ob/B.Pm (n/mm) | 12.333±5.4003 | 19.082±5.6469 | 0.05 | 17.250±5.1870 | 14.051±4.7140 | 0.29 |
| Oc.S/BS (%) | 10.601±3.6508 | 12.975±4.2334 | 0.31 | 18.439±4.7740 | 19.192±6.7888 | 0.83 |
| N.Oc/B.Pm (n/mm) | 5.3885±1.9701 | 6.3505±2.1171 | 0.42 | 8.7722±2.1935 | 9.6039±3.4546 | 0.63 |
| MS/BS (%) | 45.655±3.5366 | 43.990±4.3419 | 0.47 | 45.333±5.4009 | 44.027±1.8733 | 0.68 |
| MAR (μm/day) | 1.2265±0.2238 | 1.1889±0.1266 | 0.71 | 1.8148±0.1294 | 1.6824±0.4816 | 0.63 |
| BFR/BS (μm3/μm3/day) | 0.5634±0.1373 | 0.5220±0.0648 | 0.49 | 0.8207±0.0933 | 0.7457±0.2292 | 0.59 |
| BFR/BV (%/day) | 3.2079±1.2339 | 2.6756±0.5373 | 0.32 | 4.6232±0.4123 | 4.5078±1.7235 | 0.85 |
-
Data are means ± SD (n=6–7). BV/TV, bone volume/total volume; Tb.Th, trabecular thickness; Tb.Sp, trabecular separation; Tb.N, trabecular number; OS/BS, osteoid surface/bone surface; O.Th, osteoid thickness; Ob.S/BS, osteoblast surface/bone surface; N.Ob/B.Pm, osteoblast number/bone perimeter; Oc.S/BS, osteoclast surface/bone surface; N.Oc/BS, osteoclast number/ bone surface; N.Oc/B.Pm, osteoclast number/bone perimeter; MS/BS, mineralizing surface/ bone surface; MAR, mineral apposition ratio; BFR/BS, bone formation ratio/ one surface; BFR/BV, bone formation rate/bone surface.
Antibody list for co-IP, ChIP, and western blot.
| Antibody | Supplier | Cat Num |
|---|---|---|
| ACTIN | Santa Cruz Bio | SC47778 |
| PTH1R | Sigma | SAB4502493 |
| GATA1 | Cell Signaling Technology | 3535T |
| NFκB1 (IP) | Cell Signaling Technology | 13586S |
| NFκB1 (IB) | Cell Signaling Technology | 13681S |
| Histone H3 | Cell Signaling Technology | 9715S |
| RelB | Santa Cruz Bio | sc-48366 |
| NPAS1 | Santa Cruz Bio | sc-376083 |
| p-CREB (SER133) | Cell Signaling Technology | 9198S |
| CREB | Cell Signaling Technology | 9197T |
Primer list of ChIP-qPCR for P2-2 promoter of Pth1r.
| Prime sequence | |
|---|---|
| P2-2-Forward1 | CCATCTCTCTCACTTTCCCCAAG |
| P2-2-Reverse1 | ATCCCTGGTTCTTCGATCTAGCCC |
| P2-2- Forward2 | CCTAGCTGAACCCGAGTCTTG |
| P2-2- Reverse2 | GTCTAGCGGATCGGAGACTCT |
| P2-2- Forward3 | AACCGGGAGTCCAACGAAGGT |
| P2-2- Reverse3 | GGTCTGGCTATGTGGGGAC |
| P2-2- Forward4 | GGCTGCATAGCCTGGTTCTAGC |
| P2-2- Reverse4 | CCCACTACCCCGATCTTCCGG |
| P2-2- Forward5 | ACGGCGCGAGAAATACCAGGAG |
| P2-2- Reverse5 | CGTGGCTGGGACGTTGTCTC |