Removal of extracellular human amyloid beta aggregates by extracellular proteases in C. elegans
Figures
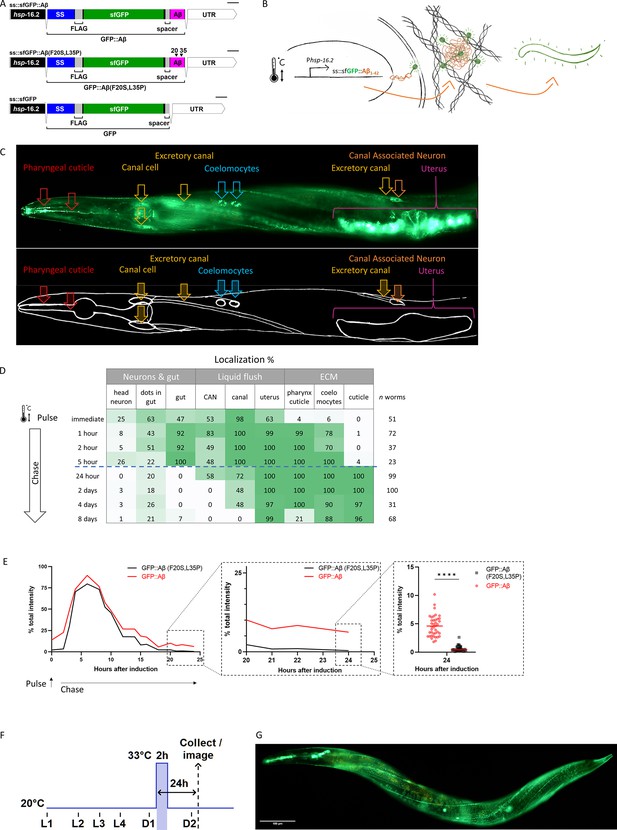
Expression of secreted human amyloid beta (Aβ) tagged with super-folder GFP (sfGFP).
(A) Genetic constructs used to generate transgenic C. elegans strains. SS indicates secretion sequence, UTR = untranslated region. (B) Hypothetical model of induction, expression, and secretion of sfGFP::Aβ in C. elegans. After a single heat-shock induction, sfGFP::Aβ is secreted and localizes to different tissues over time. (C) Localization of sfGFP::Aβ to different tissues in C. elegans. The localization to the excretory canal and the coelomocytes confirm that the sfGFP::Aβ is secreted. (D) Percentage of tissue type with sfGFP::Aβ over time. After production and secretion, most of the produced sfGFP::Aβ was flushed out, but some were retained at the cuticle up to 8 days after the induction event. (E) Clearance of sfGFP::Aβ is significantly slowed >18 hr after induction compared to non-aggregating control sfGFP::Aβ(F20S, L35P). Data represented is the average from three independent repeats combined; repeats are shown in Figure 1—figure supplement 1E. (E) sub III image is from one of the repeats, unpaired, two-tailed t-test. ****: p<0.0001. (F) Representation of methods regarding the time of heat-shock induction of expression and imaging or sample collection 24 hr after induction. (G) Representative image of the transgenic line LSD2104 and localization of secreted sfGFP::Aβ.

Validation of human amyloid beta (Aβ) present in transgenic sfGFP::Aβ C. elegans.
Day 1 wild-type (N2) and transgenic LSD2104 sfGFP::Aβ adults were heat-shocked (33°C for 2 hr) to induce Aβ aggregates. 24 hr later, when the remaining sfGFP::Aβ has formed aggregates in the cuticle, animals were harvested for protein isolation and probed with anti-Aβ peptide (MOAB-2) pan antibody, clone 6C3 (Merck #MABN254) (top panel), anti-GFP (middle panel), and monoclonal anti-α-tubulin antibody (Sigma #T9026) (bottom panel). The predicted size of the transgenic fusion protein of human Aβ1-42 with super-folder GFP (sfGFP::Aβ) is 44.6 kDa. Only after heat shock in the transgenic LSD2104 sfGFP::Aβ C. elegans, a strong band below 55 kDa and a weaker band below are visible, which correspond to the expected sizes for our transgenic fusion protein of human Aβ1-42 with sfGFP::Aβ of 44.6 kDa and cleaved one of 42.2 kDa in the anti-Aβ blot (top panel). These two bands were also present when probed with the anti-GFP in the heat shock in the transgenic LSD2104 sfGFP::Aβ C. elegans (middle panel). None of these bands were observed in wild-type nor under non-heat-shock conditions. This supports the idea that GFP aggregates contain human Aβ in the LSD2104 transgenic strain and that fluorescent sfGFP::Aβ can be used as a proxy for Aβ levels. Three independent biological trials are shown. M=marker, numbers correspond to kDa, HS = heat shock, -HS=no heat shock, N2=wild-type, sfGFP::Aβ=LSD2104.
-
Figure 1—figure supplement 1—source data 1
Full raw unedited western blots and uncropped and labeled western blots.
Raw and labeled western blots for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/83465/elife-83465-fig1-figsupp1-data1-v2.zip
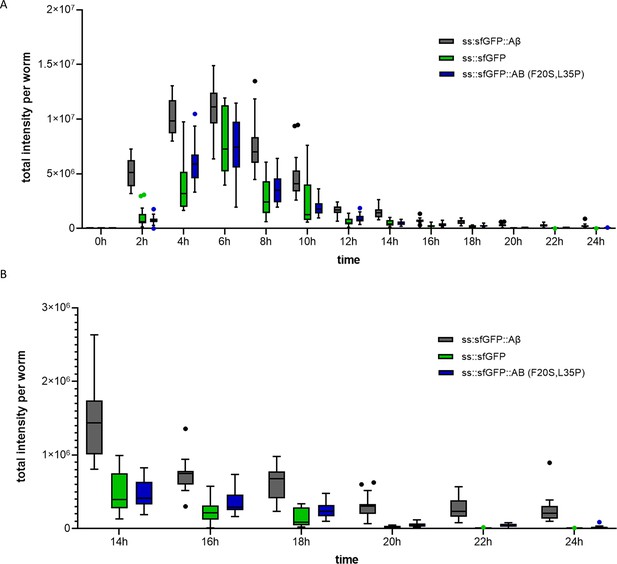
Time course of secreted amyloid beta.
(A) GFP-only (LSD1097) and non-aggregating amyloid beta (LSD1091) showed similar induction as wild-type amyloid beta. However, in contrast to wild-type amyloid beta, GFP-only, and non-aggregating amyloid beta are efficiently removed within a 24 hr timespan. (B) Tail end of A for better visibility. Plot: Tukey.

Quantification of the secreted amyloid beta time course.
Three independent trials were used to create the average for the main figure. Raw and normalized intensities are available in the data source file. (A) Independent repeats are shown together with the average used as the main figure. Each trial was normalized to its peak (100%) intensity. Sub III contains the 24 hr timepoint for each repeat. The statistical test used was an unpaired, two-tailed t-test. ****: p<0.0001 for each. (B) Raw intensity per independent trial, showing the variation within each. Plot: Tukey. (C) The last few time points for each repeat showed the sfGFP::Aβ(F20S, L35P) values are still going down, while sfGFP::Aβ seems to plateau. Plotted: Tukey.
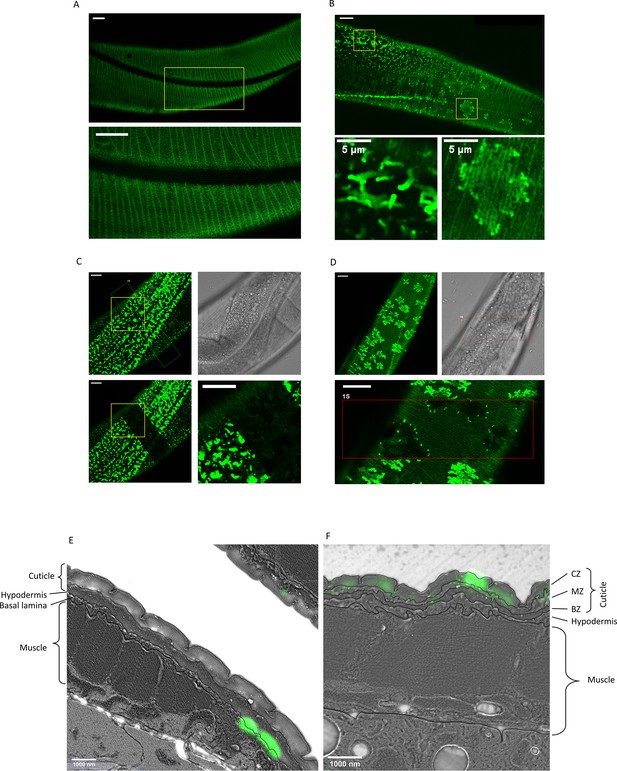
Secreted amyloid beta (Aβ) form aggregates at C. elegans extracellular matrix (ECM).
(A) The control strain sfGFP::Aβ(F20S, L35P) shows localization near the cuticle. However, the signal is relatively weak and uniform. Note. Image intensity enhanced, taken 16 hr after induction. (B) Two types of bright patterns, dubbed ‘moss’ and ‘flower’, can be observed for sfGFP::Aβ near the cuticle, 24 hr past induction of expression. (C, D) Fluorescence recovery after photobleaching shows both the moss (C) and flower (D) structures are immobile. Time of imaging up to 4 hr after bleaching. (E, F) Correlative light electron microscopy revealed localization to ECM structures. (E) Localization of the ‘moss’ structures to basal lamina when there is no muscle underneath. (F) Localization of the ‘flower’ structures to the cuticle when there is muscle underneath. CZ: cortical zone of the cuticle, MZ: medial zone of the cuticle, BZ: basal zone of the cuticle. Scale bars are 10 μm unless otherwise indicated.
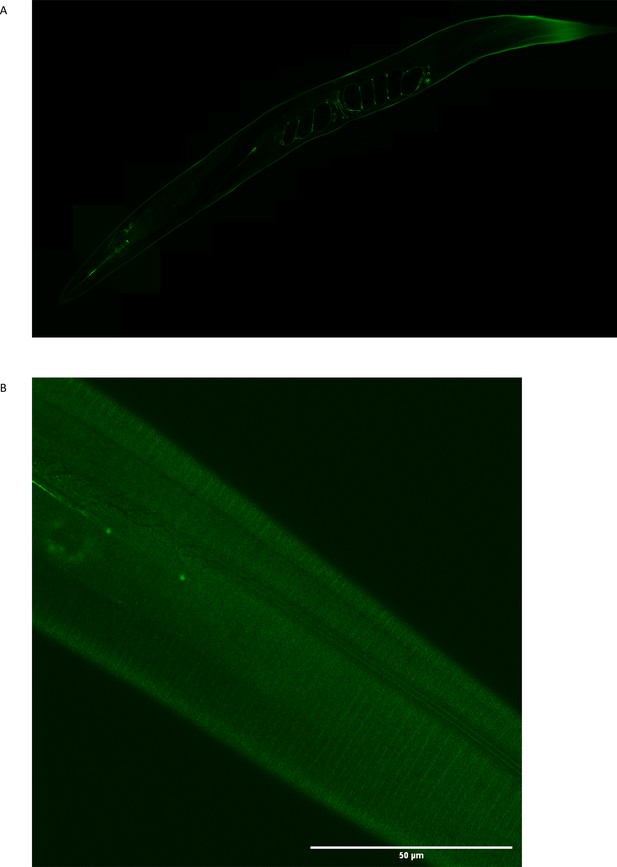
Flower and moss structures were not observed in the GFP-only strain.
The GFP-only expression appears smooth and uniform in the animal and cuticle. (A) Full-length GFP-only image, taken 16 hr after induction of expression. (B) View at the cuticle of the GFP-only strain, 16 hr after induction, brightness enhanced by 40%.
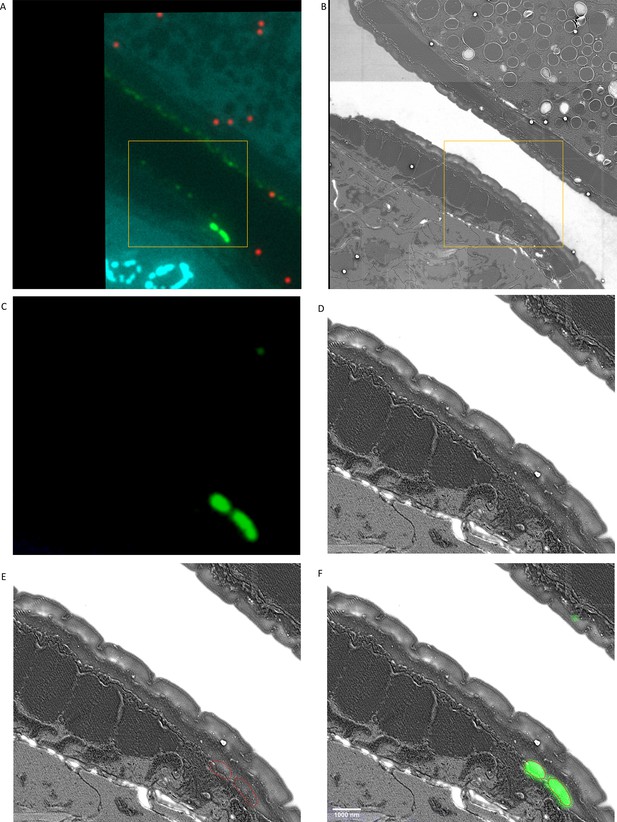
Location of sfGFP::Aβ apical to the hypodermis.
(A) Unedited fluorescence image. (B) Unedited EM image. (C) Crop of and intensity-reduced fluorescence image. (D) Corresponding crop of the EM image, contrast adjusted. (E) Overlay of EM image with the contour of GFP signal. (F) Composite image of EM and fluorescence- adjusted, cropped images. Scale bar is 1000 nm.
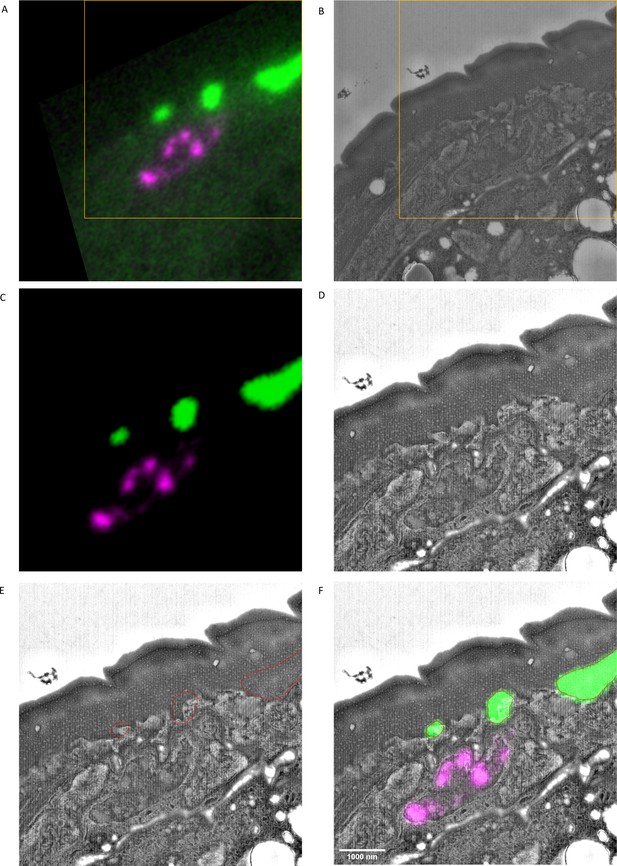
Location of sfGFP::Aβ above the hypodermis.
(A) Unedited fluorescence image. (B) Unedited EM image. (C) Crop of and intensity-reduced fluorescence image. (D) Corresponding crop of the EM image, contrast adjusted. (E) Overlay of EM image with the contour of GFP signal. (F) Composite image of EM and fluorescence- adjusted, cropped images. Scale bar is 1000 nm.

Location of sfGFP::Aβ in the cuticle.
(A) Unedited fluorescence image. (B) Unedited EM image. (C) Crop of and intensity-reduced fluorescence image. (D) Corresponding crop of the EM image, contrast adjusted. (E) Overlay of EM image with the contour of GFP signal. (F) Composite image of EM and fluorescence- adjusted, cropped images. Scale bar is 1000 nm.
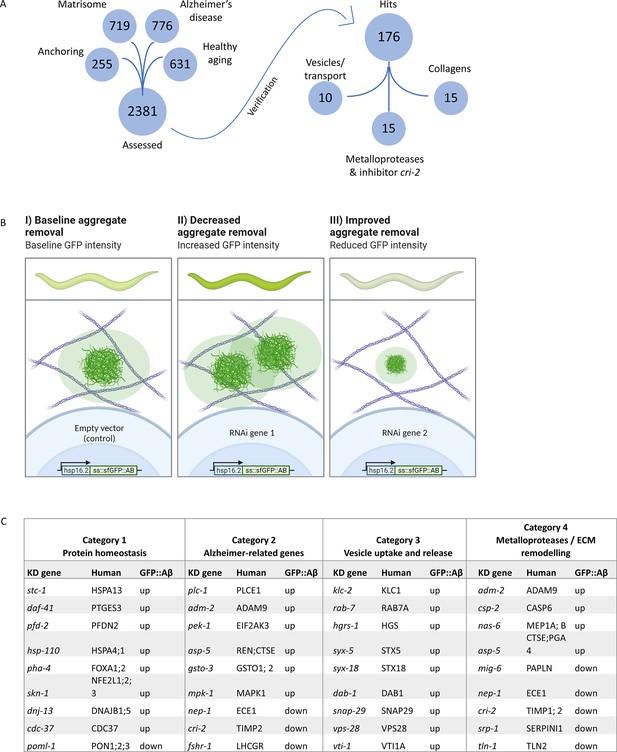
Strategy for RNA interference (RNAi) screening identified genes involved in the removal of extracellular sfGFP::Aβ.
(A) Four RNAi libraries were designed based on their hypothesized potential to affect extracellular sfGFP::Aβ aggregation. The Matrisome library contains all extracellular C. elegans genes, the Anchoring library contains transmembrane genes, the Alzheimer’s disease library is based on a meta-analysis of C. elegans orthologs of human GWAS, and the healthy aging library consists of genes that have a protective role against aging-related disease. Of the 2381 genes assessed, 176 genes were found to either increase or decrease sfGFP::Aβ load. (B) Expected fluorescence phenotypes of suppressor or enhancer genes. Grown on individual RNA clones from the L1 larval stage, populations of about 45 animals were assessed for an increase or decrease in GFP signal. An increase in signal would indicate decreased aggregate removal, while a decrease of GFP signal would imply improved aggregate removal upon knockdown of target gene expression. (C) Summary of categorization of hits shows relevant mechanisms to extracellular sfGFP::Aβ aggregate removal. Category 1 revealed screen hits in protein homeostasis, as expected when observing protein expression and turnover. Category 2 showed screen hits of orthologs of genes well known to be associated with Alzheimer’s disease in humans. Category 3 revealed screen hits of genes involved in vesicle uptake and release, processes essential to secretion and removal of extracellular proteins such as sfGFP::Aβ. Category 4 showed the involvement of metalloproteases and an inhibitor of metalloproteases. Selected candidate hits are shown; the full table and raw RNAi score are available in Supplementary file 2.
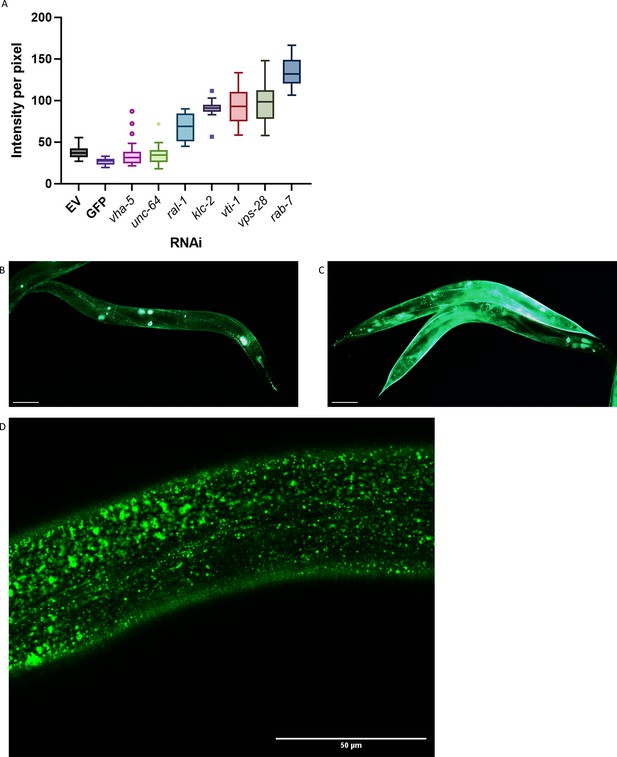
Vesicle and transport screening hits might prevent extracellular amyloid beta aggregation simply by blocking export.
(A) Intensity per pixel for all follow-up hits for the ‘vesicle’ group; genes involved in transport, secretion, or uptake of vesicle pathways. Similar results for the genes shown in A, but here, as a representative, the follow-up results are only shown for rab-7 knockdown. Knockdown by rab-7 RNAi showed high intensity of the GFP signal, yet no flower or moss aggregates at the cuticle. (B, C) RNAi of rab-7 (representative image C) showed a remarkable increase in GFP intensity compared to the empty vector (representative image B). Scale bars are 50 μm. (D) rab-7 RNAi showed a high density of vesicles near the cuticle, yet no moss or flower pattern of aggregation. Scale bar is 50 μm.
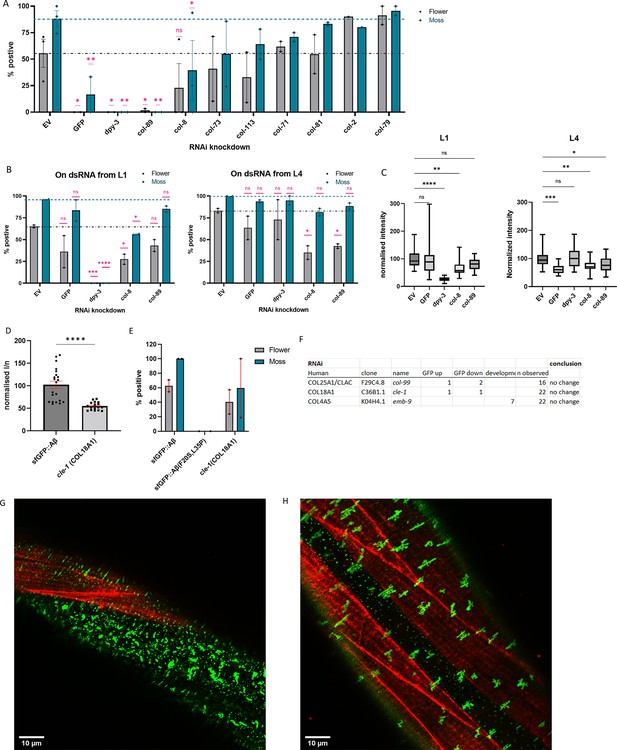
Collagens knockdown prevented or promoted extracellular sfGFP::Aβ aggregation.
(A) Collagen knockdowns that initially showed an increase in GFP intensity were followed up by observation of aggregate formation on the cuticle. RNA interference (RNAi) of collagens dpy-3 and col-89 showed no sfGFP::Aβ aggregates. Statistics: ordinary two-way ANOVA, error bars SEM. (B, C) The lack of sfGFP::Aβ aggregates could be due to a structural requirement or indirectly due to increased turnover of collagens at the extracellular matrix (ECM). To separate the two, RNAi initiated from the first larval stage (L1) was compared to RNAi initiated from the last (L4) larval stage; the latter should only take effect after the cuticle has been fully formed. (B) Score for aggregates as the % of the population. Statistics: ordinary two-way ANOVA, error bars SEM. (C) Normalized GFP intensity. Statistics: ANOVA, plotted: Tukey. (D, E) Knockout of cle-1(gk364), the ortholog of COL18A1, showed a significant reduction of sfGFP::Aβ intensity, combined with a mild reduction in flower aggregate formation. Statistics: ANOVA, error bars SEM. (F) Numbers represent individual trials for the categorized phenotype. n observed gives the total number of times the experiment was performed. RNAi of conserved collagens showed no effect on sfGFP::Aβ. (G) Colocalization of sfGFP::Aβ and collagen type IV/emb-9::mCherry showed the moss aggregates in the region of the hypodermis (absence of muscle tissue underneath). (H) Colocalization of sfGFP::Aβ and collagen type IV/emb-9::mCherry showed the flower aggregates in the region of the muscle tissue.
-
Figure 4—source data 1
Individual data values, independent trials, and statistics for Figure 4.
- https://cdn.elifesciences.org/articles/83465/elife-83465-fig4-data1-v2.xlsx

Pharmacological inhibition of collagen synthesis or cross-linking affects sfGFP::Aβ aggregate levels in the cuticle.
(A–B) Bar graphs representing reduced fluorescence intensity per pixel for sfGFP::Aβ aggregates upon treatment with collagen synthesis inhibitor BPY (A), and increased fluorescence intensity per pixel for sfGFP::Aβ aggregates upon treatment with collagen cross-linking inhibitor (B). Each dot represents a single animal. Data is plotted as a mean cumulative of three biological experiments. Error bars represent SEM. Two-tailed Welch’s t-test statistical analysis was done.
-
Figure 4—figure supplement 1—source data 1
Raw data for Figure 4—figure supplement 1.
Pharmacological inhibition of collagen synthesis or cross-linking affects sfGFP::Aβ aggregate levels in the cuticle. Individual measurements, plots, and statistics are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-fig4-figsupp1-data1-v2.xlsx
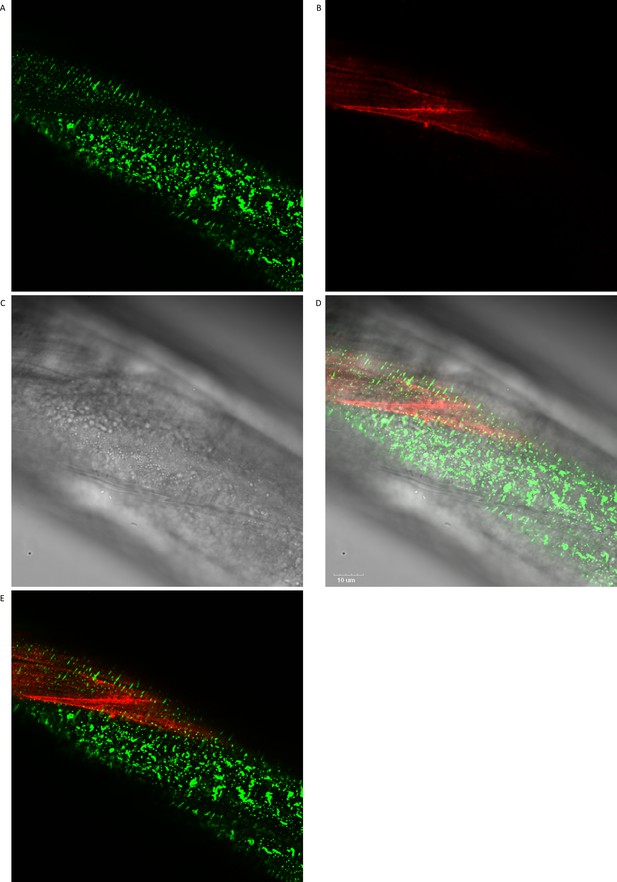
Colocalization of sfGFP::Aβ and collagen type IV/emb-9::mCherry revealed that the moss aggregates consistently colocalize to the hypodermis in the absence of muscle tissue underneath.
(A) GFP signal image. (B) mCherry signal image. (C) Normal light image. (D) All three merged images. Scale bar 10 μm. (E) GFP and mCherry signals fused.

Colocalization of sfGFP::Aβ and collagen type IV/emb-9::mCherry revealed the flower aggregates consistently colocalize above the muscle tissue.
(A) GFP signal image. (B) mCherry signal image. (C) Normal light image. (D) All three merged images. Scale bar 10 μm. (E) GFP and mCherry signals fused.
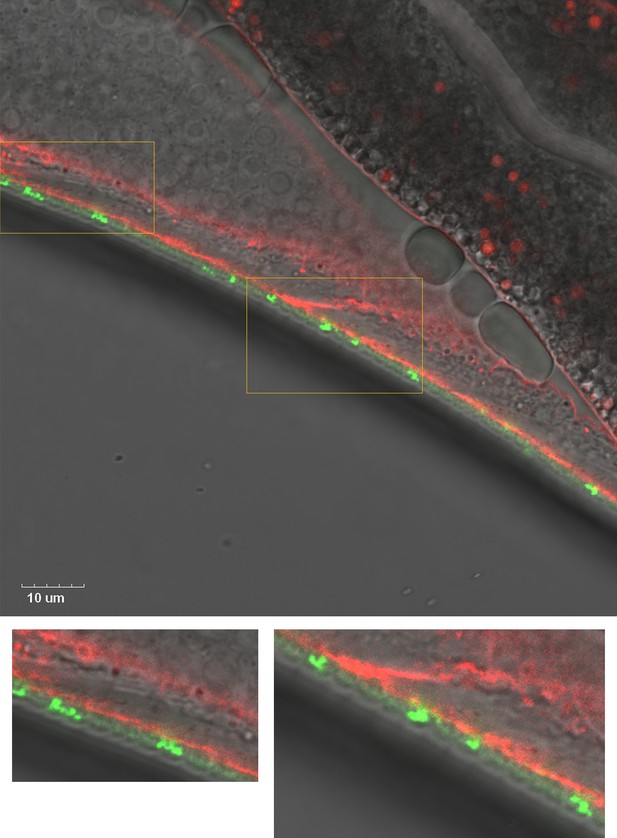
Colocalization of sfGFP::Aβ and collagen type IV/emb-9::mCherry showed that they do not colocalize.
Rather, the GFP seems to localize to the cuticle while the mCherry is situated just below the cuticle. Scale bar 10 μm.
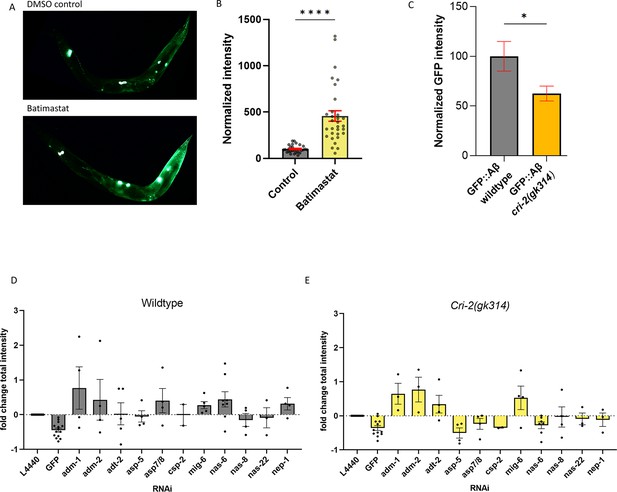
Regulation of extracellular matrix (ECM) structure and turnover influences aggregates.
(A, B) Exposure to the metalloproteinase-inhibiting drug batimastat showed a marked increase in sfGFP::Aβ, suggesting reduced removal. (A) A representative image of the GFP intensity on the DMSO control treatment and a representative image of the GFP intensity on the batimastat treatment. (B) Combined, normalized data of three independent trials. Statistics: unpaired t-test. Error bars represent SEM. (C) The deletion of the tissue inhibitor of metalloproteases (TIMP) cri-2(gk314) showed a decrease of sfGFP::Aβ load, suggesting increased activity of metalloproteases. Statistics: unpaired t-test. Error bars represent SEM. (D, E) To determine which metalloprotease under the regulation of CRI-2 showed the most potential to assist in the removal of extracellular sfGFP::Aβ, GFP intensities per population were measured for RNA interference (RNAi) of several individual metalloproteases and compared between the wild-type and cri-2 mutant background. Loss of a disintegrin and metalloprotease 2 (ADM-2) showed the largest increase in sfGFP::Aβ intensity and was selected for follow-up. Plotted: normalized mean of independent trials (each trial is one dot) with SEM.
-
Figure 5—source data 1
Quantification and raw data for Figure 5.
Individual measurements, independent biological trials, raw and processed data, and statistics are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-fig5-data1-v2.xlsx

Secreted sfGFP::Aβ intensities were similar in induction between wild-type and cri-2(gk314) backgrounds, yet, at 24 hr, less GFP signal is found for the cri-2(gk314) background.
Black line: mean. Error: SEM. (A) Total intensity measured per worm every second hour after induction up to 24 hr. Every point represents the GFP intensity of one worm. Black line: mean. Error: SEM. (B) Cropped 12–24 hr for better resolution of the tail end of the graph in A. (C) Cropped to the 24 hr timepoint for better resolution of the difference between wild-type and cri-2 mutant background. Statistics: unpaired t-test, p value 0.0201.
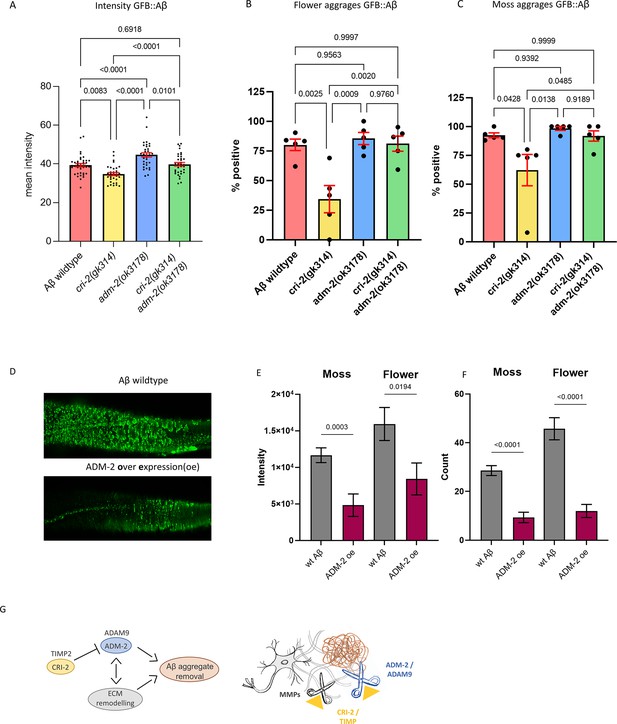
A disintegrin and metalloprotease 2 (ADM-2) was required and sufficient to remove extracellular sfGFP::Aβ.
(A) The GFP intensity for sfGFP::Aβ was reduced in the cri-2 mutant background. Loss of adm-2 increased sfGFP::Aβ intensity, even in the cri-2/adm-2 double mutant. Data were combined from four independent experiments. Statistics: ordinary two-way ANOVA. Plotted bars are mean with SEM. (B, C) Similarly, loss of cri-2 resulted in fewer animals with flower and moss aggregations, while loss of adm-2 increased both aggregation types. Loss of cri-2 and adm-2 in the double mutant background showed that other metalloproteases could not compensate for the loss of adm-2 regarding the removal of extracellular aggregates, suggesting that adm-2 was required for the removal of extracellular sfGFP::Aβ aggregates. Statistics: ANOVA. Plotted is the percentage of the population positive for aggregation type, one dot per experiment, bars are mean with SEM. (D) Visual representation of the observation that overexpression of adm-2 (ADM-2 oe) led to a reduction of extracellular sfGFP::Aβ aggregates. (E, F) Overexpression of ADM-2 was sufficient to lead to a significant reduction of sfGFP::Aβ aggregates, both in count and intensity of aggregates, measured 48 hr after induction of transgene expression. Data is the intensity and count measures over two independent experiments. Plotted are mean and SEM. Statistical analysis: unpaired t-test. (G) Schematic representation of the mechanisms in which ADM-2, under regulation by CRI-2, either directly or indirectly assisted in removing extracellular sfGFP::Aβ aggregates.
-
Figure 6—source data 1
Quantification and raw data for Figure 6.
Individual measurements, independent biological trials, raw and processed data, and statistics are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-fig6-data1-v2.xlsx

Intracellular ADM-2::mScarlet-I did not colocalize with sfGFP::Aβ.
(A, B) No colocalization of intracellular ADM-2::mScarlet-I and secreted sfGFP::Aβ aggregates. (A) Merged image of normal light, GFP, and mScarlet-I. (B) The same image as A, without the normal light, merged mScarlet-I ,and GFP signal. (C, D) No colocalization of ADM-2::mScarlet-I and sfGFP::Aβ was observed at the cuticle. (C) Merged image of normal light, GFP, and mScarlet-I. (D) The same image as C, without the normal light, merged mScarlet-I, and GFP signal. Scale bars 50 μm.

Overexpression of a disintegrin and metalloprotease 2 (ADM-2) is sufficient to lead to a significant reduction of sfGFP::Aβ aggregates.
Both the count and intensity of moss aggregates were reduced, but not of flower aggregates, when measured 24 hr after induction of ADM-2 overexpression. Data is the intensity and count measures over two independent experiments. Plotted are mean and SEM. Statistical analysis: unpaired t-test.
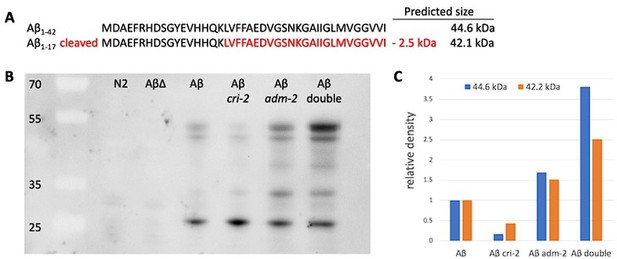
Potential direct MoA for ADM-2 in Abeta cleavage.
(A) Amino acid sequence of human Abeta with predicted cleaved-off fragment (in red) after ADAM9 cleavage. For our transgenic fusion protein of human Abeta (1-42) with super-folder GFP (sfGFP::Aβ) would result in predicted sizes of full-length sfGFP::Aβ: (B) 44.6 kDa, cleaved sfGFP::Aβ: 42.2 kDa, pure GFP: 27 kDa. (C) Western blot. Strains: wild type (N2), LSD1091 sfGFP::Aβ(F20S, L35P) non-aggregation forming (AβΔ), LSD2104 sfGFP::Aβ (Aβ), LSD2165 cri-2(gk314); sfGFP::Aβ (Aβ cri-2), LSD2201 adm-2(ok1204); sfGFP::Aβ (Aβ adm-2), LSD2204 adm-2(ok1204); cri-2(gk314); sfGFP::Aβ (Aβ double). Antibody: 1st anti-GFP (mouse)2nd anti-mouse-HRP. Sample preparation adapted from Groh et al., 2017 doi: 10.3791/56464: 1. Collect worm pellet and lyse with tissue lyser according to lab protocol. 2. Centrifuge at 18,400 x g for 20 min at 4 °C. Collect the supernatant. 3. Resuspend the pellet in 100 µL RIPA buffer and centrifuge at 18,400 x g for 20 min at 4°C. Discard the supernatant, spin shortly, and be careful to remove leftover supernatant. 4. To solubilize the highly insoluble proteins, resuspend the pellet in 75 µL Urea/SDS buffer (8 M urea, 2% SDS, 50 mM dithiothreitol (DTT), 50 mM Tris, pH 8) and incubate for 10 min at room temperature. (C) Relative density. The LSD2104 sfGFP::Aβ (Aβ) was taken as the normal reference point to build the ratio of full-length sfGFP::Aβ (44.6 kDa) vs cleaved sfGFP::Aβ (42.2 kDa).
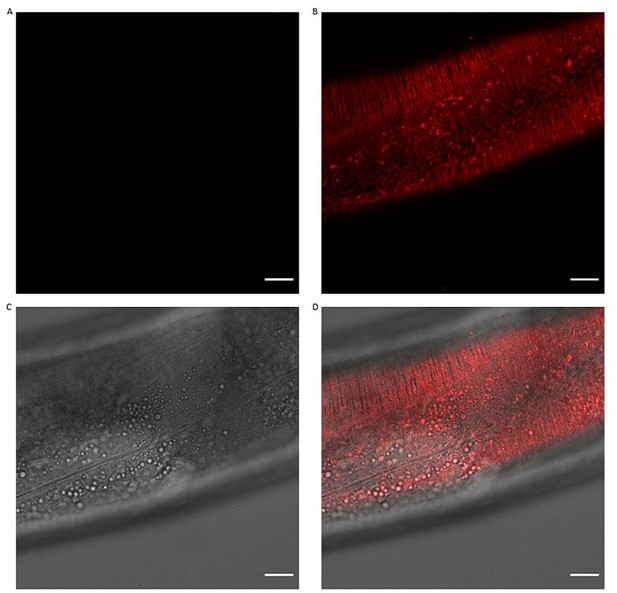
Collagen ROL-6::mScarlet.
Representative image of ROL-6::mScarlet pattern both underneath the muscle and hypodermal tissues. A) GFP channel B) mScarlet-I channel C) DCI D) combined. Scale bars 10μm.
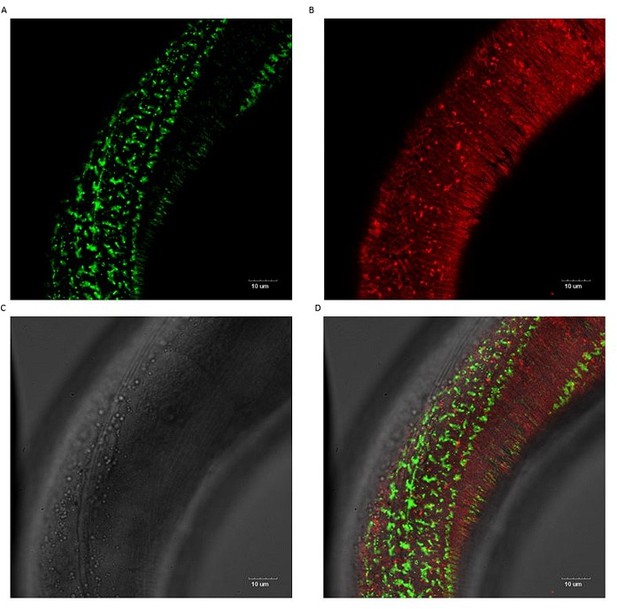
No co-localization of sfGFP::Aβ and collagen ROL-6::mScarlet.
No co-localization is observed. The cuticle collagen ROL-6 tagged with wrmScarlet-I (codon-optimized mScarlet fluorophore) showed no obvious difference in pattern nor brightness between wildtype and Aβ expressing strains. Scale bars 10μm.
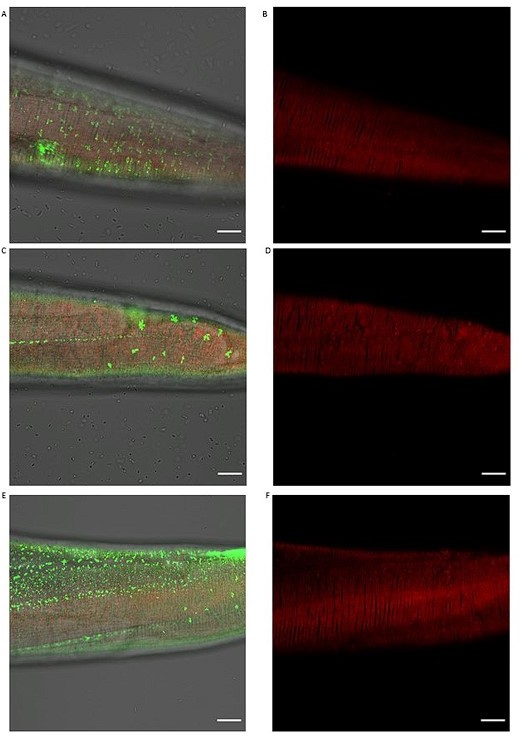
Knockdown of different cuticular collagens affects sfGFP::Aβ aggregates but not the ECM morphology represented by collagen ROL-6::mScarlet.
(A,B) Knockdown of col-119 by RNAi resulted in a slightly lower number of sfGFP::Aβ but no obvious effect on the collagen structure of ROL-6::mScarlet. (C,D) Knockdown of col-136 by RNAi resulted in a lower number of sfGFP::Aβ but no obvious effect on the collagen structure of ROL-6::mScarlet. (E, F) Knockdown of col-137 by RNAi resulted in a high number of smaller sfGFP::Aβ aggregates with very few bigger sfGFP::Aβ aggregates but no obvious effect on the collagen structure of ROL-6::mScarlet. Scale bars 10μm.
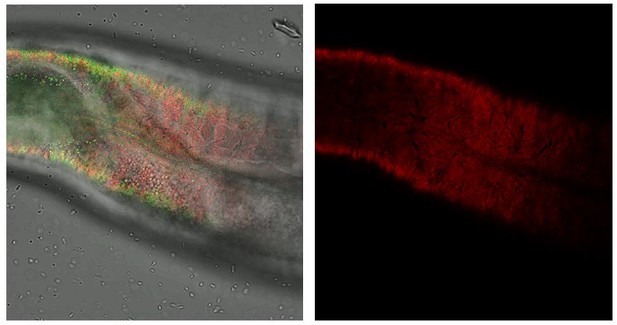
Knockdown of cuticular collagen dpy-7 affects sfGFP::Aβ aggregates and the ECM morphology represented by collagen ROL-6::mScarlet.
Knockdown of dpy-7 by RNAi resulted in the absence of sfGFP::Aβ but also effect on the collagen structure of ROL-6::mScarlet. Scale bars 10μm.
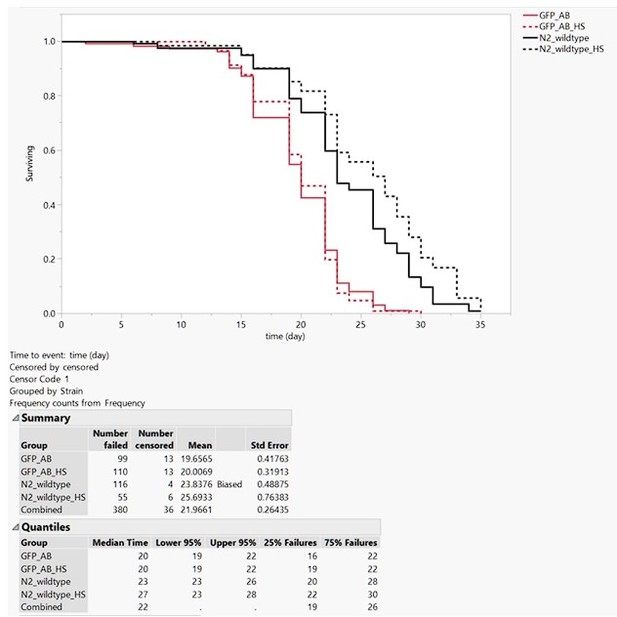
Lifespan assay.
At day one of adulthood, wild type (N2) and LSD2104 (GFP_AB) were heat shocked for 2h at 33°C and placed on 50 µM FUdR NGM plates at 20°C to assess their lifespan (Protocol as in Ewald et al., 2016 https://doi.org/10.1111/acel.12509). The GFP_AB (LSD2104 sfGFP::Aβ) strain was shorter-lived as wild type N2. This could be due to the inherent leakiness at 20°C of hsp-16 promoter driving the sfGFP::Aβ expression (as reported for other transgenes and lifespan in Ewald et al., 2016 https://doi.org/10.1111/acel.12509). However, the single heat shock, which induced the sfGFP::Aβ, did not further shorten the lifespan of the transgenic LSD2104 strain. We know that the heat shock had worked due to the slight increase in lifespan of the heat-shock wild type (a phenomenon also reported in Ewald et al., 2016 https://doi.org/10.1111/acel.12509).

Embryonic survival assay.
Upon heat shock, sfGFP::Aβ is expressed in eggs. We aimed to determine the effects of sfGFP::Aβ on embryonic survival and hatching into the first larval stage. 10 N2 or LSD2104 (ss::sfGFP::Aβ) gravid adults were allowed to lay eggs for 3h. Then either heat shocked at 33°C for 3h (HS) or not (No HS). The number of hatched vs non-hatched eggs was scored 24 h later. There was no significant difference under non- or heat-shocked conditions of embryonic survival of LSD2104 vs N2. Note that the y-axis of the graph starts at 80%..
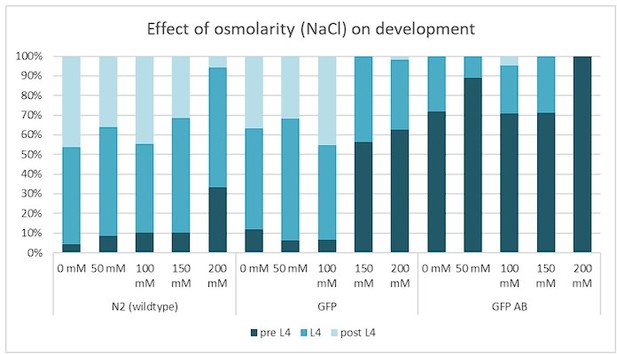
Development and proteotoxicity stress assay.
The rationale of this assay was to determine whether perturbing protein homeostasis via increasing osmolarity would affect larval development. Wild type (N2), GFP-only LSD1091 [Phsp-16.2::ss::GFP] (GFP), and LSD2104 sfGFP::Aβ (GFP AB) adults were heat shocked for 2h at 33°C and F1 eggs were placed on NGM plates containing 0, 50, 100, 200 mM NaCl and the larval stage was scored 48h at 25°C. For N2 only at 200 mM salt, the development was slowed, whereas sfGFP::Aβ (GFP AB) already had a bit slower development, which was further slowed at 200 mM salt. Given that the difference pre-L4 to L4 is a few hours and rather minor, this suggests that sfGFP::Aβ (GFP AB) had minor proteotoxicity effects on development.

Cuticle integrity assay.
Since sfGFP::Aβ forms aggregates in the cuticle (ECM), we sought to determine whether that would affect cuticle barrier function. LSD2104 sfGFP::Aβ adults were heat shocked for 2h at 33°C (HS) or not (no HS) and then placed in low bleach concentration and M9 to determine the time until cuticle breaks. Three biological repeats (1,2,3) are shown that are all non-significant. (Experimental details: 500 µL NaCl + 1mL NaOH (1M) were added to a total volume of 10 mL in M9. A 24-well plate was used to add 1 mL of this bleach solution per well. In each well, 15 worms were placed, at which time point a timer was started. Scored was the time point for each worm cuticle to break by visual observation with a dissecting scope.).

Touch habituation.
Previously, we had shown that overexpression of APL-1 (the homolog of the APP) showed impairment in touch habituation (Ewald et al., 2012, DOI: https://doi.org/10.1523/JNEUROSCI.0495-12.2012). To determine whether sfGFP::Aβ also affects learning plasticity, we compared N2 wild type (WT) with LSD2104 sfGFP::Aβ. Day 1 adults were heat shocked for 3h at 33°C, and 24h later, the responses to touch with a hair lash to the head and tail were recorded. The total time was recorded until the animal became unresponsive (A; data plotted as a bar graph). Also, the total number of touches until the animal became unresponsive was counted, and data was plotted as a bar graph (B). The bar graph shown here represents the cumulative mean data for two independent trials. Error bars are shown as SEM. Each dot represents a single animal. 2-way Anova was performed for statistical analysis. Transgenic LSD2104 sfGFP::Aβ animals had higher resistance towards the touch and took longer time to become habituated to touch as compared to wildtype even under non-heat shocked conditions (i.e., when sfGFP::Aβ was not induced). Heat shock-induced aggregate formation, however, had no effect on touch habituation. This result suggests that Aβ aggregate formation in the extracellular matrix does not affect tactile learning plasticity.

Chemotaxis assay.
Previously, we had shown that overexpression of APL-1 (the homolog of the APP) showed impairment in olfactory and gustatory learning behavior (Ewald et al., 2012, DOI: https://doi.org/10.1523/JNEUROSCI.0495-12.2012). To determine whether sfGFP::Aβ also affects olfactory learning plasticity, we compared N2 wild type (WT) with LSD2104 sfGFP::Aβ. Day-1 adults were heat shocked for 3h at 33°C, and 24h later, a chemotaxis assay was performed. Benzaldehyde was used as the test chemical, while ethanol served as the control. Briefly, a 90 mm unseeded NGM containing petri plate was divided into four quadrants, and a dot was placed equidistant from the center inside each of the quadrants. 1 µL ethanol was spotted in two diagonally opposite quadrants and benzaldehyde in the other two. 1:200 was the dilution used for benzaldehyde (0.005%). 1 µL of 1M Sodium azide was also pipetted in all the four spots. Approx 100-200 animals were exposed to the chemicals by placing them in a 1 cm circle marked in the center. For pre-conditioning, animals were placed in the center and exposed to 0.6 µL of undiluted benzaldehyde by placing the chunks of agar containing the benzaldehyde on the lid of the Petri plate. After pipetting the animals, let the water evaporate before closing the lid. The animals were incubated with benzaldehyde for one hour at 20 ℃. Pre-conditioned animals were assayed for chemotaxis behavior after one hour of incubation with benzaldehyde. After one hour, plates were shifted to 4 ℃ to freeze the animals in their position. One hour later, animals were counted in each quadrant and the center. The Chemotaxis Index (C.I.) was calculated as (the number of animals in test – the number of animals in control) divided by the total number of animals in the plate. Bar graph represents the chemotaxis index as the mean value of three independent trials for each condition. Error bars are SEM. 2-way Anova was performed for statistical analysis. We observed that sfGFP::Aβ transgenic animals had lower C.I. as compared to wild-type, which was not affected by heat-induced aggregate formation. Pre-conditioned animals, as expected, showed reduced C.I. in all conditions. This result suggests that aggregate formation did not affect the chemotaxis behavior towards benzaldehyde in neither naive nor pre-conditioned environments.

Head region of sfGFP::Aβ and collagen type IV/emb-9::mCherry.
We performed confocal imaging of the head region of sfGFP::Aβ and collagen type IV/emb-9::mCherry. By the bulb outlined by the basement membrane of emb-9::mCherry (in red), there are several cells in green (i.e., sfGFP::Aβ) that could be the nerve ring and neurons.

Pan-neuronal expressing sfGFP::Aβ C. elegans arrest in L1.
Shown is a GFP channel image above and the same image in the bright field below. GFP-positive animals are arrested in L1 (green tiny worms), and all the adult worms visible in the bright field image are GFP-negative, suggesting that neuronal expressing sfGFP::Aβ C. elegans arrest in L1.
Additional files
-
Supplementary file 1
RNA interference (RNAi) assessed genes and libraries.
Individual measurements, independent biological trials, raw and processed data, and statistics are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-supp1-v2.xlsx
-
Supplementary file 2
RNA interference (RNAi) hits and categories.
Individual measurements, independent biological trials, raw and processed data, and statistics are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-supp2-v2.xlsx
-
Supplementary file 3
Ortholog collagens data overview.
Detailed overview about the collagens orthologs.
- https://cdn.elifesciences.org/articles/83465/elife-83465-supp3-v2.xlsx
-
Supplementary file 4
Reagents list.
Detailed lists of C. elegans strains, bacterial strains, RNA interference (RNAi) clones, plasmids, and primers are provided.
- https://cdn.elifesciences.org/articles/83465/elife-83465-supp4-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83465/elife-83465-mdarchecklist1-v2.docx





