Regulatory and coding sequences of TRNP1 co-evolve with brain size and cortical folding in mammals
Figures
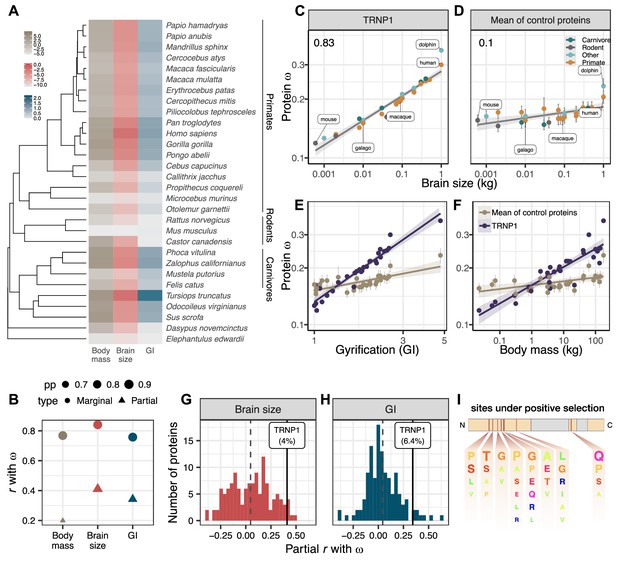
TRNP1 amino acid substitution rates co-evolve with brain size and cortical folding in mammals.
(A) Mammalian species for which body mass, brain size, gyrification index (GI) measurements, and TRNP1 coding sequences were available (n=30)(Figure 1—figure supplement 1). Log2-transformed units: body mass and brain size in kg; GI is a ratio (cortical surface/perimeter of the brain surface). (B) Estimated marginal and partial correlation between ω of TRNP1 and the three traits using Coevol (Lartillot and Poujol, 2011). Size indicates posterior probability (pp). (C) TRNP1 protein substitution rates (ω) significantly correlate with brain size (, = 0.97).(D) The average correlation across 124 control proteins with brain size (=0.10). (E) TRNP1 ω correlation with GI compared to the average across control proteins. (F) TRNP1 ω correlation with body mass compared to the average across control proteins. (C, D, E, F) Error bars indicate standard errors. (G) Distribution of partial correlations between ω and brain size of the control proteins and TRNP1. (H) Distribution of partial correlations between ω and GI of the control proteins and TRNP1. (I) Scheme of the mouse TRNP1 protein (223 amino acids [AAs]) with intrinsically disordered regions (orange) and sites (red lines) subject to positive selection in mammals (ω > 1, Figure 1—figure supplement 1). Letter size of the depicted AAs represents the abundance of AAs at the positively selected sites.
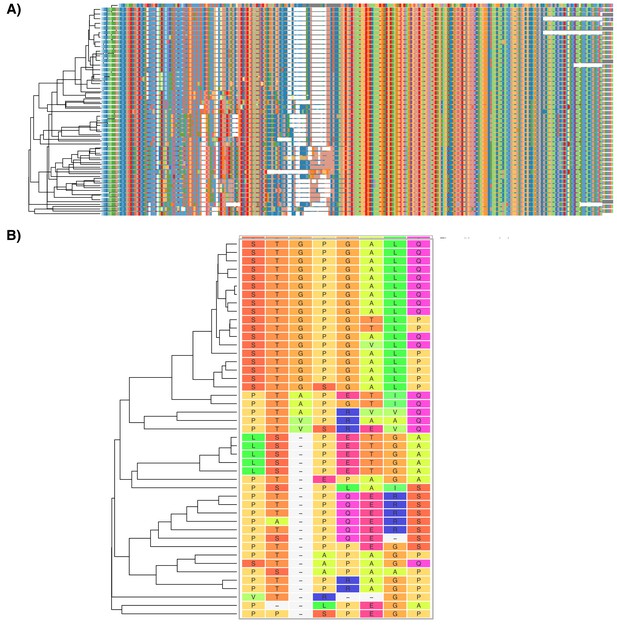
TRNP1 protein-coding sequence analysis.
(A) Multiple alignment of 45 TRNP1-coding sequences (99.0% completeness) using phylogeny-aware aligner PRANK. The alignment is 735 bases long, which translates to 245 amino acids (AAs). For comparison: human TRNP1 coding sequence is 227 AA long, whereas mouse – 223 AAs. (B) Sites under positive selection across the phylogenetic tree according to PAML M8 site model (in total 9.8% of sites with ω>1, likelihood ratio test (LRT), p-value < 0.001). The depicted sites had a posterior probability Pr(ω > 1) > 0.95 according to naive empirical Bayes analysis. Colours of the amino acids indicate their relatedness in biochemical properties. Sites with light-grey background and a dash indicate indels.
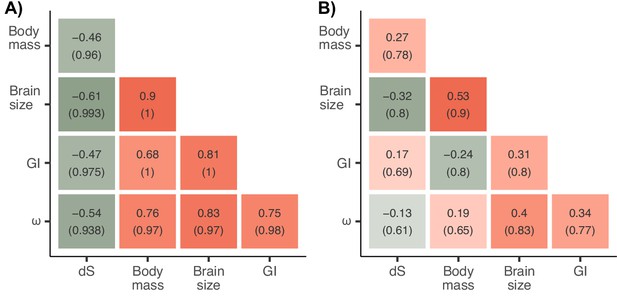
Estimated marginal (A) and partial (B) correlation matrices of the combined Coevol model including the three traits and substitution rates of TRNP1.
Posterior probabilities of the associations are depicted in brackets.
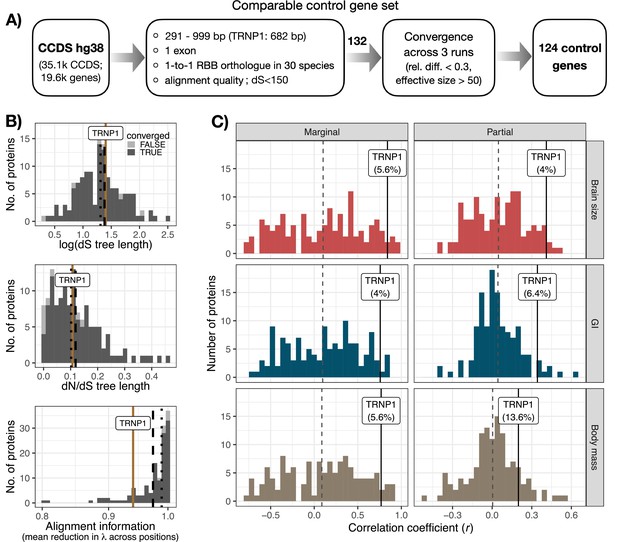
Control protein evolution rate correlation with brain size, gyrification, and body mass.
(A) A flowchart depicting the selection of comparable control proteins to infer the average protein correlation rate with the included phenotypes across the mammalian phylogeny of 30 species. 1088 1-exon protein sequences with a comparable length to TRNP1 were available in human CCDS, 132 of which had good full-length alignment quality across the 30 species, a log(dS) tree length less than 3× SD away from the average and all belong to a different gene. 124 of these converged between the three Coevol runs. RBB – reciprocal best blat. (B) TRNP1 and 132 control proteins show comparable synonymous substitution rates (log(dS), top) and protein evolution rates (ω, middle), inferred using PAML branch free-ratios model. All included proteins have high-quality full-length alignments (bottom), quantified as the mean relative tree length across all alignment positions per protein. Brown lines indicate TRNP1, black dashed lines – the average across all proteins, black dotted lines – median across all proteins. (C) Marginal and partial correlation distribution of 125 proteins, including TRNP1, with the three phenotypes: brain size, gyrification index (GI), and body mass inferred using Coevol. Dashed lines indicate the average across all proteins.
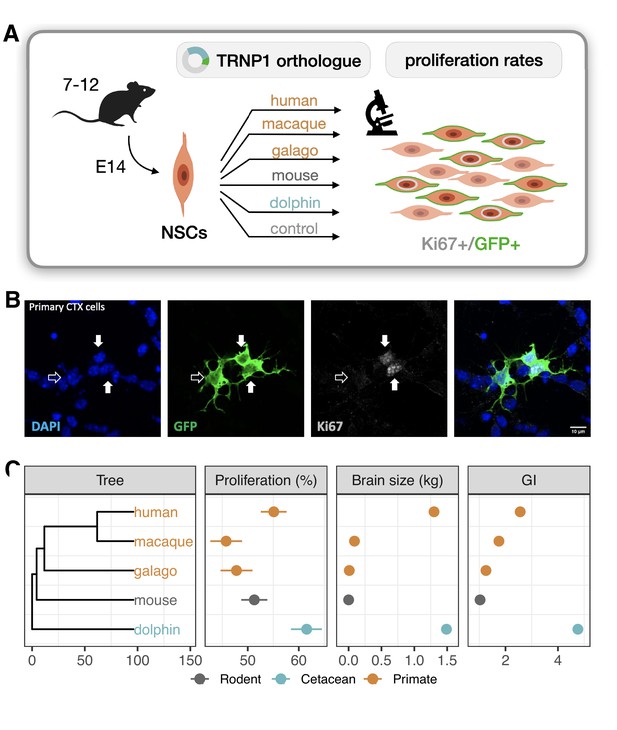
TRNP1 proliferative activity correlates with brain size and cortical folding.
(A) Five different TRNP1 orthologues were transfected into neural stem cells (NSCs) isolated from cerebral cortices of 14-day-old mouse embryos and proliferation rates were assessed after 48 hr using Ki67 immunostaining as proliferation marker and green fluorescent protein (GFP) as transfection marker in 7–12 independent biological replicates. (B) Representative image of the transfected cortical NSCs immunostained for GFP and Ki67. Arrows indicate three transfected cells of which two (solid arrows) are Ki67-positive (Figure 2—figure supplement 1). (C) Induced proliferation in NSCs transfected with TRNP1 orthologues from five different species (Supplementary file 2). Proliferation rates are a significant predictor for brain size (=10.04, df = 1, BH-adjusted p-value = 0.0018 = 11.75 ± 2.412, = 0.89) and GI (=5.85, df = 1, BH-adjusted p-value = 0.016 = 16.97 ± 6.568, = 0.69) in the respective species (phylogenetic generalized least squares [PGLS], likelihood ratio test [LRT]). Error bars indicate standard errors. Included species: human (Homo sapiens), rhesus macaque (Macaca mulatta), northern greater galago (Otolemur garnettii), house mouse (Mus musculus), common bottlenose dolphin (Tursiops truncatus).
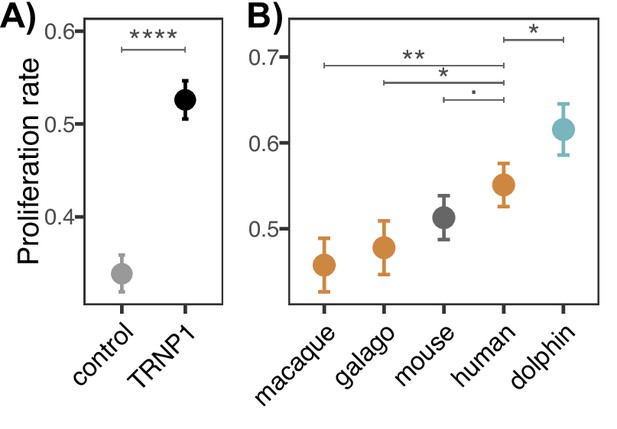
Proliferation induced by TRNP1.
(A) Proliferation induced in neural stem cells (NSCs) transfected with TRNP1 (all TRNP1 orthologues combined) compared to the control NSCs transfected only with green fluorescent protein (GFP). TRNP1 presence in NSCs significantly increases the proliferation rates (TRNP1: 0.53 (±0.02), control: 0.34 (±0.02), df = 57). (B) Proliferation induced in NSCs transfected with TRNP1 orthologous from five different species (Supplementary file 2). (A), (B) Bars indicate standard errors of logistic regression and asterisks indicate the significance of pairwise comparisons (Tukey test, p-value: <0.1, *<0.05, **<0.001, ****<2e-16).
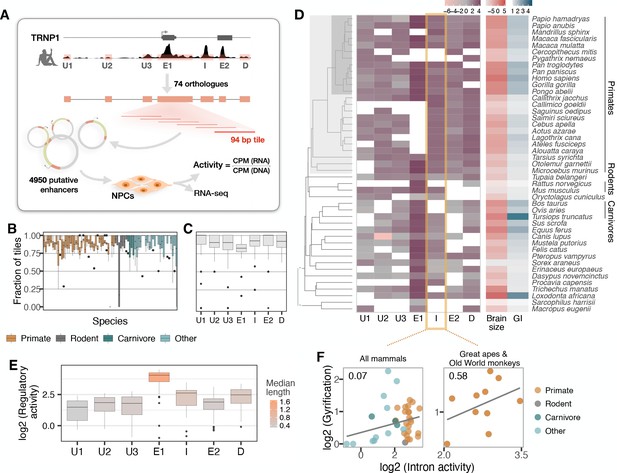
Activity of a cis-regulatory element (CRE) of TRNP1 correlates with cortical folding in catarrhines.
(A) Experimental setup of the massively parallel reporter assay (MPRA). Regulatory activity of seven putative TRNP1 CREs from 75 species were assayed in neural progenitor cells (NPCs) derived from human and cynomolgus macaque induced pluripotent stem cells. (Figure 3—figure supplement 1). (B) Fraction of the detected CRE tiles in the plasmid library per species across regions. The detection rates are unbiased and uniformly distributed across species and clades with only one extreme outlier Dipodomys ordii. (C) Fraction of the detected CRE tiles in the plasmid library per region across species. (D) Log-transformed total regulatory activity per CRE in human NPCs across species with available brain size and gyrification index (GI) measurements (n=45). (E) Total activity per CRE across species. Exon 1 (E1), intron (I), and the downstream (D) regions are more active and longer than other regions. (B, C, E) Each box represents the median and first and third quartiles with the whiskers indicating the furthest value no further than 1.5 * IQR from the box. Individual points indicate outliers. Figure 3—figure supplement 2 (F) Regulatory activity of the intron CRE is weakly associated with gyrification across mammals (phylogenetic generalized least squares [PGLS], likelihood ratio test [LRT] p-value=0.097, R2=0.07, n=37) and strongest across great apes and Old World monkeys, that is, catarrhines (PGLS, LRT p-value=0.003, R2=0.58, n=10).

Length of the covered cis-regulatory element (CRE) sequences in the massively parallel reporter assay (MPRA) library across the tree.
Species for which the regions were inferred based on DNase hypersensitive sites (DHS) from embryonic brain (Bernstein et al., 2010) are marked in bold and black: human (H. sapiens) and mouse (M. musculus). These species do not show extreme differences in length compared to others (human: 5/7, mouse: 3/5 regions within the 10% and 90% quantiles). The orthologous CRE sequence length differs strongly between primate and non-primate species, being on average 1.8–2.8 times longer in the primate species than in the other mammals.
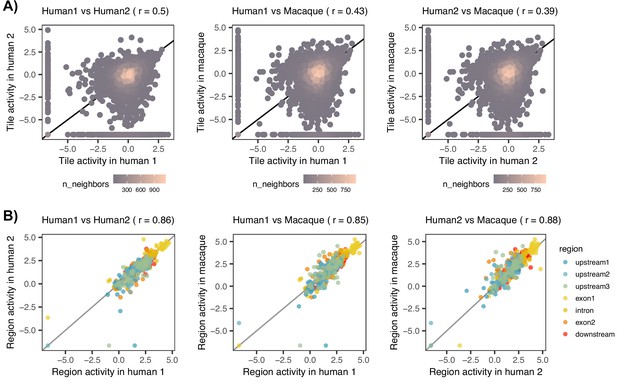
Analysis of massively parallel reporter assay (MPRA) data.
(A) Pairwise correlation of the log2-transformed cis-regulatory element (CRE) tile activity between the three transduced cell lines: human 1, human 2, and macaque. Pearson’s is specified in the brackets of figure titles. (B) Pairwise correlation of the log2-transformed summarized activity per CRE region between cell lines. Pearson’s is specified in the brackets of figure titles.
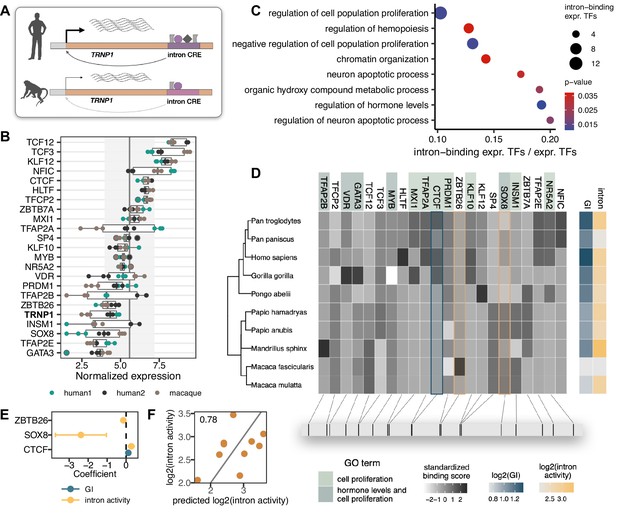
Transcription factors (TFs) with binding site enrichment on intron cis-regulatory elements (CREs) regulate cell proliferation and are candidates to explain the observed activity across catarrhines.
(A) Orthologous intron CRE sequences show different regulatory activities under the same cellular conditions, suggesting variation in cis regulation across species. (B) Variance-stabilized expression in neural progenitor cells (NPCs) of TRNP1 and the 22 TFs with enriched binding sites (motif weight ≥ 1) on the intron CREs. Each box represents the median, first and third quartiles with the whiskers indicating the furthest value no further than 1.5 * IQR from the box. Points indicate individual expression values. Vertical line indicates average expression across all 392 TFs (5.58), grey area: standard deviation (1.61). (C) Eight top enriched biological processes (Gene Ontology, Fisher’s exact test p-value <0.05) of the 22 TFs. Background: all expressed TFs (392). (D) Variation in binding scores of the enriched TFs across catarrhines. Heatmaps indicate standardized binding scores (grey), gyrification index (GI) values (blue) and intron CRE activities (yellow) from the respective species. TF background colour indicates gene ontology assignment of the TFs to the two most significant biological processes. The bottom panel indicates the spatial position of the top binding site (motif score >3) for each TF on the human sequence. (E) Binding scores of three TFs (CTCF, ZBTB26, SOX8) are the best candidates to explain intron CRE activity, whereas only CTCF binding shows an association with the GI (phylogenetic generalized least squares [PGLS], likelihood ratio test [LRT] p-value <0.05). (F) Predicted intron CRE activity by the binding scores of the three TFs vs. the measured intron CRE activity across catarrhines.
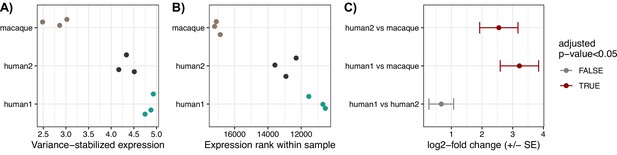
TRNP1 expression in human and cynomolgus macaque (Macaca fascicularis) cell lines.
(A) Variance-stabilized expression of TRNP1. (B) Expression ranks of TRNP1. (C) Differential expression results between the three cell lines. Colour indicates significant differential expression using Benjamini-Hochberg adjusted p-value cutoff of 0.05.
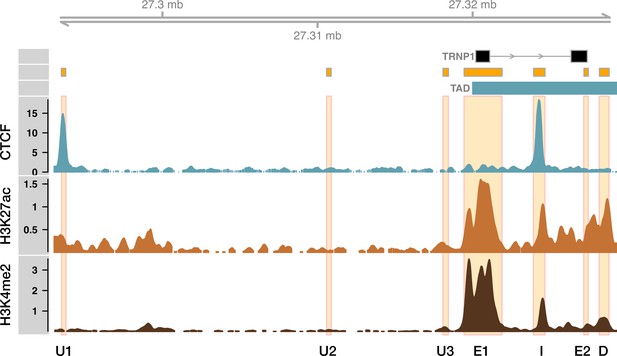
Human genome tracks for the TRNP1 locus (hg19).
TAD boundary (blue) ends just upstream of the TRNP1 exon 1 in human germinal zone (gestational week 8, Hi-C) (Won et al., 2016). In agreement, CTCF ChIP-seq peak was detected within intron (I) CRE (neural progenitor cells [NPCs]). Another nearby CTCF binding site that is bound by CTCF is in the upstream 1 (U1) CRE sequence (ENCODE Project Consortium (2012)). Histonemark H3K27ac (light brown) and H3K4me2 (dark brown) data from human developing cortex (gestational week 7, Reilly et al., 2015).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (45 mammal species) | TRNP1 | See Supplementary file 1a | See Supplementary file 1a | See Supplementary file 1a |
| Strain, strain background (E. coli) | NEB 10-beta | New England Biolabs; Rowley, MA, United States | Cat# C3020K | Electrocompetent E. coli |
| Strain, strain background (E. coli) | NEB 5-alpha High Efficiency | New England Biolabs; Rowley, MA, United States | Cat# C2987I | Chemically competent E. coli |
| Cell line (Macaca fascicularis) | Cynomolgus Macaque NPC | This paper, based on Geuder et al., 2021 | N15_39B2 | Macaca fascicularis neural progenitor cells |
| Cell line (Mus musculus) | N2A | ATCC; Manassas, VA, United States | CCL-131 | |
| Cell line (Homo sapiens) | HEK293T | ATCC; Manassas, VA, United States | CRL-11268 | |
| Cell line (Homo sapiens, female) | Human NPC 1 | This paper, based on Geuder et al., 2021 | N4_29B5 | Human neural progenitor cells |
| Cell line (Homo sapiens, male) | Human NPC 2 | This paper, based on Geuder et al., 2021 | N4_12 C2 | Human neural progenitor cells |
| Biological sample (Mus musculus) | Primary murine cerebral cortex cells (NSC) | This paper, based on Esgleas et al., 2020 | primary | See Methods |
| Sequence-based reagent | MPRA oligo Library Trnp1 CRE | Custom Array; Redmond, WA, United States | custo | See https://github.com/Hellmann-Lab/Co-evolution-TRNP1-and-GI |
| Transfected construct (multiple species) | MPRA Library in lentiviral particles | This paper | custom | Lentiviral particles with pMPRA-lenti and TRNP1 CRE library |
| Antibody | rabbit anti Ki67 (monoclonal) | Abcam; Waltham, MA, United States | Cat# ab92742, Clone EPR3610 | 1:100 |
| Antibody | chicken anti-GFP (polyclonal) | Aves Labs; Davis, CA, United States | RRID: AB_2307313, Cat# GFP-1010, Polyclonal | 1:500 |
| Recombinant DNA reagent | pCAG-GFP_Gateway plasmid | Dr. Paolo Malatesta | NA | Kind gift of Dr. Paolo Malatesta |
| Recombinant DNA reagent | pMDLg/pRRE plasmid | Addgene; Waterton, MA, United States | Addgene 12251 | |
| Recombinant DNA reagent | pRSV-Rev plasmid | Addgene; Waterton, MA, United States | Addgene 12253 | |
| Recombinant DNA reagent | pMD2.G plasmid | Addgene; Waterton, MA, United States | Addgene 12259 | |
| Recombinant DNA reagent | pMPRAlenti1 plasmid | Addgene; Waterton, MA, United States | Addgene 61600 | Kind gift of Dr. Davide Cacchiarelli |
| Recombinant DNA reagent | pNL3.1[Nluc/minP] plasmid, SfiI restriction site mutated | Dr. Davide Cacchiarelli | NA | Kind gift of Dr. Davide Cacchiarelli |
| Recombinant DNA reagent | pMPRA1 plasmid | Addgene; Waterton, MA, United States | Addgene 49349 | Kind gift of Dr. Davide Cacchiarelli |
| Recombinant DNA reagent | pENTR1a plasmid | Stahl et al., 2013 | pENTR1a | |
| Peptide, recombinant protein | hEGF | Miltenyi Biotec; Bergisch Gladbach, Germany | Cat#130-093-825 | |
| Peptide, recombinant protein | B-27 Supplement | Thermo Fisher Scientific; Waltham, MA, United States | Cat#12587–010 | |
| Peptide, recombinant protein | N2 Supplement | Thermo Fisher Scientific; Waltham, MA, United States | Cat#17502048 | |
| Peptide, recombinant protein | L-Ascorbic acid 2-phosphate | Sigma/Merck; St. Louis, MO, United States | Cat#A8960-5G | |
| Peptide, recombinant protein | poly-D-lysine | Sigma/Merck; St. Louis, MO, United States | Cat# A-003-E | |
| Peptide, recombinant protein | bFGF | PeproTech, Cranbury, New Jersey, United States | Cat#100-18B | |
| Commercial assay or kit | GenomiPhi V2 DNA-Amplification Kit | Sigma/Merck; St. Louis, MO, United States | Cat# GE25-6600-32 | |
| Commercial assay or kit | Gateway LR Clonase Enzyme mix | Thermo Fisher Scientific; Waltham, MA, United States | Cat# 11791019 | |
| Commercial assay or kit | Lipofectamine 2000 | Thermo Fisher Scientific; Waltham, MA, United States | Cat# 11668019 | |
| Commercial assay or kit | Lipofectamine 3000 | Thermo Fisher Scientific; Waltham, MA, United States | Cat# L3000015 | |
| Commercial assay or kit | Micellula DNA Emulsion & Purification Kit | Roboklon; Berlin, Germany | Cat# E3600-01 | |
| Commercial assay or kit | Agilent High Sensitivity DNA Kit | Agilent; Santa Clara, CA, United States | Cat# 5067–4626 | |
| Commercial assay or kit | Nextera XT DNA Library Preparation Kit | Illumina; San Diego, CA, United States | Cat# FC-131–1024 | |
| Chemical compound, drug | GlutaMax-I | Thermo Fisher Scientific; Waltham, MA, United States | Cat# 35050038 | |
| Chemical compound, drug | Blasticidin S HCl | Thermo Fisher Scientific; Waltham, MA, United States | Cat# R21001 | |
| Chemical compound, drug | DMEM-GlutaMAX | Thermo Fisher Scientific; Waltham, MA, United States | Cat# 10566016 | |
| Chemical compound, drug | Polybrene | Sigma/Merck; St. Louis, MO, United States | Cat# TR-1003-G | |
| Chemical compound, drug | TRI reagent | Sigma/Merck; St. Louis, MO, United States | Cat# T9424-200ML | |
| Chemical compound, drug | Geltrex | Thermo Fisher Scientific; Waltham, MA, United States | Cat# A1413302 | |
| Sequence-based reagent | Trnp1 CRE resequencing primers | Integrated DNA Technologies, Coralville, IO, United States | custom | See https://github.com/Hellmann-Lab/Co-evolution-TRNP1-and-GI |
| Sequence-based reagent | Trnp1 coding resequencing forward primer | Integrated DNA Technologies, Coralville, IO, United States | custom | GGGAGGAGTAAACACGAGCC |
| Sequence-based reagent | Trnp1 coding resequencing reverse primer | Integrated DNA Technologies, Coralville, IO, United States | custom | AGCCAGGTCATTCACAGTGG |
| Software, algorithm | Hotspot version 4.0.0 | John et al., 2011, http://www.uwencode.org/software/hotspot | NA | |
| Software, algorithm | BLAT version 35x1 | Kent, 2002, https://github.com/djhshih/blat | NA | |
| Software, algorithm | PriMux, compiled on 20 July 2014 | Hysom et al., 2012, https://sourceforge.net/projects/primux/ | NA | |
| Software, algorithm | deML version 1.1.3 | Renaud et al., 2015, https://github.com/grenaud/deml | NA | |
| Software, algorithm | cutadapt version 1.6 | Martin, 2011, https://anaconda.org/bioconda/cutadapt | NA | |
| Software, algorithm | Trinity version 2.0.6 | Grabherr et al., 2011, https://github.com/trinityrnaseq/trinityrnaseq/releases | NA | |
| Software, algorithm | rBLAST version 0.99.2 | https://github.com/mhahsler/rBLAST | NA | |
| Software, algorithm | PRANK version 150803 | Löytynoja, 2021, http://wasabiapp.org/software/prank/ | NA | |
| Software, algorithm | PAML version 4.8 | Yang, 1997, http://abacus.gene.ucl.ac.uk/software/paml.html | NA | |
| Software, algorithm | Coevol version 1.4 | Lartillot and Poujol, 2011, https://megasun.bch.umontreal.ca/People/lartillot/www/downloadcoevol.html | NA | |
| Software, algorithm | NextGenMap (NGM) version 0.0.1 | Sedlazeck et al., 2013, http://cibiv.github.io/NextGenMap/ | NA | |
| Software, algorithm | Primer Blast | Ye et al., 2012 | NA | |
| Software, algorithm | zUMIs version 2.4.5b | Parekh et al., 2018, https://github.com/sdparekh/zUMIs | NA | |
| Software, algorithm | STAR version STAR_2.6.1 c | Dobin et al., 2013, https://github.com/alexdobin/STAR | NA | |
| Software, algorithm | DESeq2 version 1.26.0 | Love et al., 2014, Bioconductor | NA | |
| Software, algorithm | Cluster Buster, compiled on Jun 13 2019 | Frith et al., 2003, http://cagt.bu.edu/page/ClusterBuster_download | NA | |
| Software, algorithm | R version 3.6/4 | https://www.r-project.org/ | NA | |
| Software, algorithm | nlme version 3.1–143 | https://cran.r-project.org/web/packages/nlme/index.html | NA | |
| Software, algorithm | topGO version 2.40.0 | Alexa, 2009, https://bioconductor.org/packages/release/bioc/html/topGO.html | NA | |
| Software, algorithm | ape version 5.4 | https://cran.r-project.org/web/packages/ape/index.html | NA | |
| Software, algorithm | multcomp version 1.4–13 | https://cran.r-project.org/web/packages/multcomp/index.html | NA | |
| Software, algorithm | RR2 version 1.0.2 | https://cran.r-project.org/web/packages/rr2/index.html | NA |
Additional files
-
Supplementary file 1
Summaries of all information for the Coevol analyses, including the data sources for genome sequence and phenotype information as well as relevant Coevol outputs.
Source information on TRNP1 protein sequences (1a), primate gDNA (1b), phenotype information (1c) as well as detailed results from PAML (Yang, 1997) (1d) and Coevol (Lartillot and Poujol, 2011) results for TRNP1 and the control proteins (1f, 1e, 1g).
- https://cdn.elifesciences.org/articles/83593/elife-83593-supp1-v1.xlsx
-
Supplementary file 2
Model selection for NSC proliferation (2a) as well as proliferation rates based on the selected model (2b) and statistical testing of pairwise differences (2c).
- https://cdn.elifesciences.org/articles/83593/elife-83593-supp2-v1.xlsx
-
Supplementary file 3
Analyses of TRNP1 CREs and their activities and a characterization of TF binding sites within.
TRNP1 DNase hypersensitive sites (3a), phylogenetic generalized least squares (PGLS) model selections using likelihood ratio test for all seven CREs and the whole phylogeny (3b) as well as only the intron CRE in Old World monkeys and great apes (3c) and enriched gene ontologies based on the transcription factors (TFs) with binding site enrichment in the intron CRE (3d).
- https://cdn.elifesciences.org/articles/83593/elife-83593-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83593/elife-83593-mdarchecklist1-v1.pdf





