Interactions between metabolism and growth can determine the co-existence of Staphylococcus aureus and Pseudomonas aeruginosa
Figures
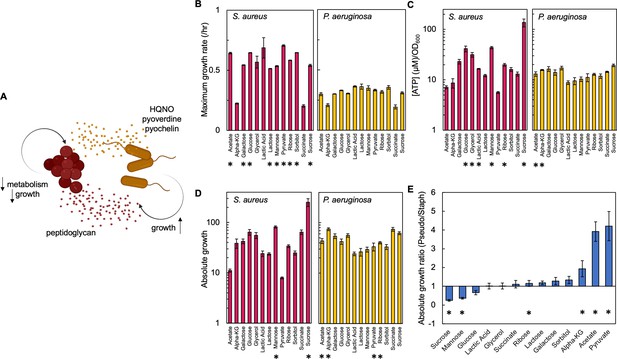
Carbon source in the growth medium affects growth and metabolism in P. aeruginosa and S. aureus; together this determines absolute growth.
(A) Core interactions that affect growth and metabolism of S. aureus (red) and P. aeruginosa (yellow) when in co-culture. Peptidoglycan from S. aureus activates the expression of virulence factors from P. aeruginosa. These virulence factors, pyoverdine, pyochelin and HQNO, reduce metabolism and growth of S. aureus, which provides a benefit to P. aeruginosa. (B) Maximum growth rate of S. aureus (left) and P. aeruginosa (right) when grown in TSB medium with different carbon sources. Growth curves in Figure 1—figure supplement 1. Average from a minimum of three biological replicates. Error bars are standard error of the mean (SEM). * indicates a significantly greater maximum growth rate (two-tailed t-test, P≤0.027). For panels B, C, and D the exact number of biological replicates and all p values are in Supplementary file 1. (C) The concentration of ATP (μM) produced by S. aureus (left) and P. aeruginosa (right) grown in TSB medium with different carbon sources. SEM from a minimum of four biological replicates each consisting of three technical replicates. * indicates a significantly greater concentration of ATP (two-tailed t-test, p≤0.030). (D) Absolute growth of S. aureus (left) and P. aeruginosa (right) grown in TSB medium with different carbon sources. Absolute growth calculated using data shown in panels B and C. SEM from a minimum of three biological replicates. * indicates a significant difference in absolute growth between both species (two-tailed t-test, p≤0.046). (E) Ratio of absolute growth between P. aeruginosa and S. aureus. Data from panel D. * indicates a significant difference as determined using data in panel D.
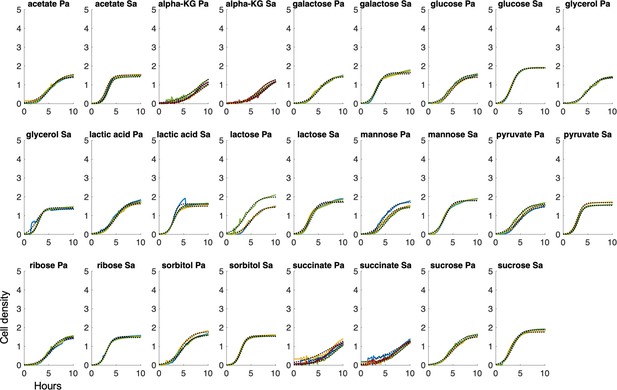
Determining growth rate.
Raw growth curves for S. aureus and P. aeruginosa in TSB medium with various carbon sources. Black line = fit from a logistic growth curve. Different colored lines = individual biological replicates. Sa = S. aureus, Pa = P. aeruginosa. alpha-KG = α-ketoglutarate. Cell density and time are plotted consistently on each axis.
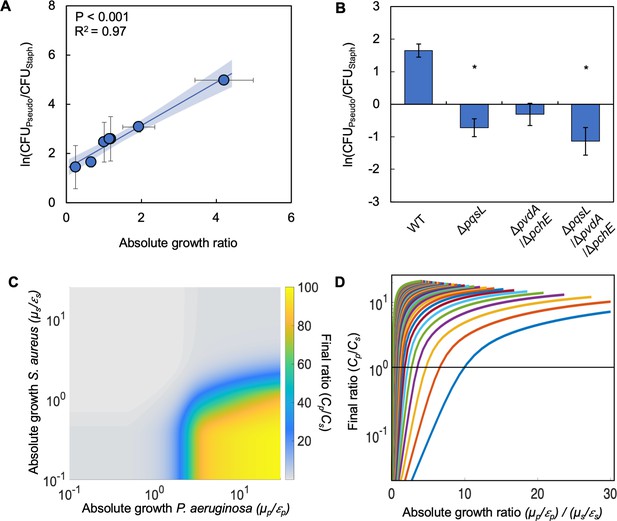
Differences in absolute growth determine the final densities of S. aureus and P. aeruginosa in co-culture.
(A) Final density ratio of P. aeruginosa to S. aureus after 24 hr of growth in co-culture with different carbon sources affording different absolute growth ratios. Standard error of the mean (SEM) from a minimum of four biological replicates. R2 and P values shown on plot are from a linear regression. Weighted least squares regression: R2=0.99, p<0.0001. Kruskal-Wallis, p=0.0151 (Shapiro-Wilk<0.0001). For panels A and B, final cell density in CFU for both strains shown in Figure 2—figure supplement 1. Linear regressions between the final density ratio and [ATP], maximum growth rates, the ratio of [ATP], and the ratio of growth rates shown in Figure 2—figure supplement 1. For panels A and B, exact number of biological replicates and all P values shown in Supplementary file 2. Shaded region indicates 95% confidence interval. (B) Final density ratio of P. aeruginosa knockout strains to S. aureus after 24 hr of growth in co-culture. Carbon source included in the growth medium was glucose. SEM from five biological replicates. * indicates significantly different from the wildtype final density ratio (Dunn test with wildtype ratio as control group; p≤0.0309; Kruskal-Wallis; p=0.0051; Shapiro-Wilk<0.001). (C) Heat map showing the effect of absolute growth (μ/ε) of P. aeruginosa (Cp) and S. aureus (Cs) on the final ratio of the strains. For these simulations, growth rate (μ) was held constant while metabolism (ε) was varied. For simulations where growth rate is varied and metabolism is held constant, see Figure 2—figure supplement 2. For panels C and D, simulations were performed using Equations 1–3. Total simulation time = 24 hr. Parameters in Supplementary file 2. Model description and development in Methods. Sensitivity analysis in Supplementary file 2. (D) Representative simulations showing the relationship between the ratio of absolute growth and the final density ratio of P. aeruginosa to S. aureus. For these simulations, growth rate (μ) and metabolism (ε) for P. aeruginosa were varied while they were fixed for S. aureus. Each colored line presents a combination of μ and ε for P. aeruginosa. Simulations using fixed values for P. aeruginosa and varied values for S. aureus shown in Figure 2—figure supplement 2.

Raw density (CFU/mL) of P. aeruginosa and S. aureus in co-culture in TSB medium.
(A) Raw bacterial density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in TSB medium containing various carbon sources. This raw data was used to produce Figure 2A. Standard error of the mean (SEM) from a minimum of three biological replicates. Alpha-KG = α-ketoglutarate. p Values and the exact number of biological replicates can be found in Supplementary file 2. (B) Final density ratio of P. aeruginosa to S. aureus plotted as a function of [ATP] produced by P. aeruginosa when in monoculture. In panels B-G, SEM from a minimum of three biological replicates; R2 and p values on plot are from a linear regression. All final density ratios replotted from Figure 2A. All growth rates replotted from Figure 1B. All ATP concentrations replotted from Figure 1C. (C) Final density ratio of P. aeruginosa to S. aureus plotted as a function of [ATP] produced by S. aureus when in monoculture. SEM from a minimum of three biological replicates. (D) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of P. aeruginosa when in monoculture. SEM from a minimum of three biological replicates. (E) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of S. aureus when in monoculture. SEM from a minimum of three biological replicates. (F) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of maximum growth rates (P. aeruginosa/S. aureus). SEM from a minimum of three biological replicates. (G) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of [ATP] (P. aeruginosa/S. aureus). SEM from a minimum of three biological replicates. (H) Left: Final density ratio of P. aeruginosa to S. aureus after 24 hr of growth in TSB medium with different carbon sources. TSB medium was overlaid with 3 mL of mineral oil. Values above each bar represent the absolute growth ratio. p=0.013, Kruskal-Wallis (Shapiro-Wilk, p=0.0084). * indicates a significant difference in the density of S. aureus and P. aeruginosa (Mann-Whitney, p=0.0013; Shapiro Wilk, p<0.0001). Right: Raw density (CFU/mL) of P. aeruginosa and S. aureus used to determine final density ratio in the left panel. SEM from a minimum of three biological replicates. All p values and exact number of biological replicates can be found in Supplementary file 3. (I) Raw density (CFU/mL) of S. aureus and knockout strains of P. aeruginosa in co-culture in TSB medium containing glucose. This raw data was used to produce Figure 2B. (J) Bacterial density at OD600=0.1 – the initial OD600 that experiments are standardize to. p=0.37, two-tailed t-test. Values above each bar are the average OD600 values from which bacterial density was measured. p=0.73, two-tailed t-test. SEM from six biological replicates. (K) Average maximum growth rate of wildtype and knockout strains of P. aeruginosa grown in TSB medium with different carbon sources. SEM from at least three biological replicates. ANOVA ≥0.0814 with the exception of lactic acid (p=0.0167). Dunnett’s test with wildtype as the control (ΔpqsL, P≥0.1911; ΔpvdA/ΔpchE, P≥0.3952; ΔpqsL/ΔpvdA/ΔpchE, p≥0.1656). * indicates significantly different (P=0.0429). See Supplementary file 2 for exact number of biological replicates and all P values. (L) Raw growth curves of P. aeruginosa knockout strains grown in TSB medium with various carbon sources. Black line = fit from a logistic growth curve. Different colored lines = individual biological replicates. alpha-KG = α-ketoglutarate. Cell density and time are plotted consistently on each axis.
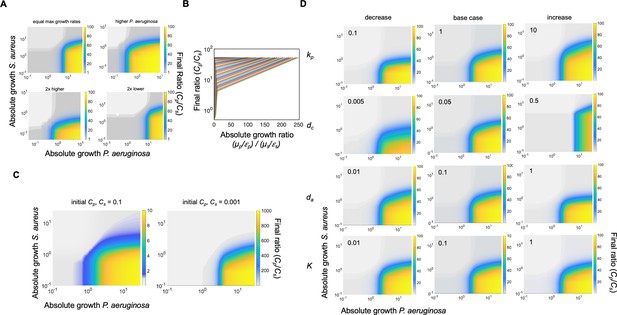
Additional simulations and sensitivity analysis using our mathematical model (Equations 1–3).
(A) Heat map showing the effect of absolute growth (μ/ε) of P. aeruginosa (Cp) and S. aureus (Cs) on the final ratio of the species. For these simulations, the value of metabolism (ε) for both species ranged over the same parameter spaces as in Figure 2C. While all other parameters stayed constant (Supplementary file 2), we varied the growth rates of P. aeruginosa and S. aureus. Top left: equal growth rates of both species, μp=μs = 0.2. Top right: growth rate of P.aeruginosa is higher than S. aureus (μp = 0.8, μss0.4). Bottom left: both growth rates doubled from base case (μp = 0.64, μss0.8). Bottom right: both growth rates reduced by half from base case (μp = 0.16, μss0.2). For all panels, total simulation time = 24 hours. Model description and development in Methods. (B) Simulations showing the relationship between the absolute growth ratio and the final density ratio of P. aeruginosa to S. aureus. For these simulations, growth rate (μ) and metabolism (ε) for S. aureus were varied while they were fixed for P. aeruginosa. Each colored line represents a combination of μ and ε for S. aureus. (C) Simulations showing the effect of reducing the initial density of the population. Simulations were performed as in Figure 2C. (D) Sensitivity analysis. Each heat map shows the effect of changing the value of the indicated variable (Equations 1–3) 10-fold relative to the base case (center column of panel, base case parameter set in Supplementary file 2). Variable being increased or decreased shown on the left side of each row. For these simulations, growth rate (μ) was held constants while metabolism (ε) was varied. Total simulation time = 24 hr.
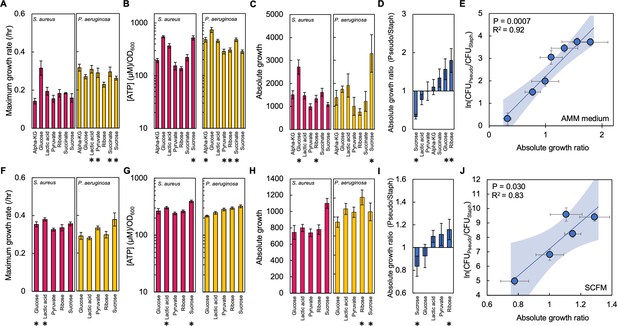
The absolute growth ratio predicts community composition in additional growth media.
(A) Maximum growth rate of S. aureus (left) and P. aeruginosa (right) when grown in AMM medium with different carbon sources. Growth curves in Figure 3—figure supplement 1. Standard error of the mean (SEM) from a minimum of eight biological replicates. * indicates a significantly greater maximum growth rate (two-tailed t-test, P≤0.0033). For all panels in this figure, the exact number of biological replicates and all p values are in Supplementary file 4. (B) The concentration of ATP (μM) produced by S. aureus (left) and P. aeruginosa (right) grown in AMM medium with different carbon sources. Concentration of ATP determined using a standard curve. SEM from six biological replicates each consisting of two technical replicates. * indicates a significantly greater concentration of ATP (two-tailed t-test, P≤0.011). (C) Absolute growth of S. aureus (left) and P. aeruginosa (right) grown in AMM medium with different carbon sources. Absolute growth calculated using data shown in panels A and B. * indicates a significant difference in absolute growth between both species (two-tailed t-test, p≤0.03). SEM from a minimum of six biological replicates. (D) Ratio of absolute growth between P. aeruginosa and S. aureus when grown in AMM medium. Data from panel C. * indicates a significant difference as determined using data in panel C. (E) Final density ratio of P. aeruginosa to S. aureus after 24 hr of growth in co-culture in AMM medium with different carbon sources affording different absolute growth ratios. SEM from a minimum of five biological replicates. R2 and p values shown on plot are from a linear regression. Weighted least squares regression: R2=0.93, p=0.0005. Kruskal-Wallis, p<0.0001 (Shapiro-Wilk=0.0001). Final cell density in CFU/mL in Figure 3—figure supplement 2. Linear regressions between the final density ratio and [ATP], maximum growth rates, the ratio of [ATP], and the ratio of growth rates shown in Figure 3—figure supplement 2. Shaded region indicates 95% confidence interval. (F) Maximum growth rate of S. aureus (left) and P. aeruginosa (right) when grown in SCFM with different carbon sources. Growth curves in Figure 3—figure supplement 1. SEM from a minimum of five biological replicates. * indicates a significantly greater maximum growth rate (two-tailed t-test, p≤0.035). (G) The concentration of ATP (μM) produced by S. aureus (left) and P. aeruginosa (right) grown in SCFM with different carbon sources. SEM from five biological replicates each consisting of two technical replicates. * indicates a significantly greater concentration of ATP (two-tailed t-test, p≤0.04). (H) Absolute growth of S. aureus (left) and P. aeruginosa (right) grown in SCFM with different carbon sources. Absolute growth calculated using data shown in panels F and G. * indicates a significant difference in absolute growth between both species (two-tailed t-test, p≤0.04). (I) Ratio of absolute growth between P. aeruginosa and S. aureus when grown in SCFM. Data from panel H. * indicates a significant difference as determined using data in panel H. (J) Final density ratio of P. aeruginosa to S. aureus after 24 hr of growth in co-culture in SCFM with different carbon sources affording different absolute growth ratios. SEM from a minimum of four biological replicates. R2 and p values shown on plot are from a linear regression. Weighted least squares regression: R2=0.91, p=0.0128. Kruskal-Wallis, p<0.0001 (Shapiro-Wilk <0.0001). Final cell density in CFU/mL shown in Figure 3—figure supplement 2. Linear regressions between the final density ratio and [ATP], maximum growth rates, the ratio of [ATP], and the ratio of growth rates shown in Figure 2. Shaded region indicates 95% confidence interval.

Raw growth curves of bacteria grown in either (A) AMM medium or (B) SCFM.
Black line = fit from a logistic growth curve. Different colored lines = individual biological replicates. Sa = S. aureus, Pa = P. aeruginosa. alpha-KG = α-ketoglutarate. Cell density and time are plotted consistently on each axis.

Raw density (CFU/mL) of P. aeruginosa and S. aureus in co-culture in AMM medium and SCFM.
(A) Raw bacterial density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in AMM medium containing various carbon sources. This raw data was used to produce Figure 3E. Standard error of the mean (SEM) from a minimum of five biological replicates. For all panels in the figure, p values and exact number of biological replicates can be found in Supplementary file 4. (B) Final density ratio of P. aeruginosa to S. aureus plotted as a function of [ATP] produced by P. aeruginosa when in monoculture in AMM medium. In panels B-G, SEM from a minimum of five biological replicates; R2 and p values on plot are from a linear regression. All final density ratios replotted from Figure 3E. All growth rates replotted from Figure 3A. All ATP concentrations replotted from Figure 3B. SEM from a minimum of five biological replicates. (C) Final density ratio of P. aeruginosa to S. aureus plotted as a function of [ATP] produced by S. aureus when in monoculture (AMM medium). SEM from a minimum of five biological replicates. (D) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of P. aeruginosa when in monoculture (AMM medium). SEM from a minimum of five biological replicates. (E) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of S. aureus when in monoculture (AMM medium). SEM from a minimum of five biological replicates. (F) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of [ATP] (P. aeruginosa/S. aureus) in AMM medium. SEM from a minimum of five biological replicates. (G) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of maximum growth rates (P. aeruginosa/S. aureus) in AMM medium. SEM from a minimum of five biological replicates. (H) Raw bacterial density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in SCFM containing various carbon sources. This raw data was used to produce Figure 3J. SEM from a minimum of four biological replicates. (I) Final density ratio of P. aeruginosa to S. aureus plotted as a function of [ATP] produced by P. aeruginosa when in monoculture in SCFM. In panels I-N, SEM from a minimum of four biological replicates; R2 and P values on plot are from a linear regression. All final density ratios replotted from Figure 3H. All growth rates replotted from Figure 3F. All ATP concentrations replotted from Figure 3G. SEM from a minimum of four biological replicates. (J) Final density ratio of P. aeruginosa to S. aureus plotted as a function of ATP produced by S. aureus when in monoculture (SCFM). SEM from a minimum of four biological replicates. (K) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of P. aeruginosa when in monoculture (SCFM). SEM from a minimum of four biological replicates. (L) Final density ratio of P. aeruginosa to S. aureus plotted as a function of maximum growth rate of S. aureus when in monoculture (SCFM). SEM from a minimum of four biological replicates. (M) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of [ATP] (P. aeruginosa/S. aureus) in SCFM. SEM from a minimum of four biological replicates. (N) Final density ratio of P. aeruginosa to S. aureus plotted as a function of the ratio of maximum growth rates (P. aeruginosa/S. aureus) in SCFM. SEM from a minimum of four biological replicates.
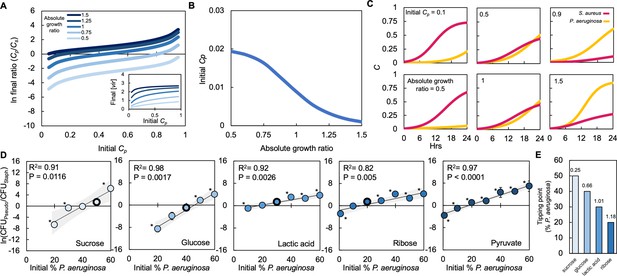
Absolute growth determines tipping point of co-existence of P. aeruginosa and S. aureus in co-culture.
(A) Simulations showing the effect of changing the initial fraction of P. aeruginosa (Cp) and S. aureus (Cs) on the final density ratio. Each colored line represents simulations performed with different absolute growth values (as indicated in the figure legend). For panels A-C, simulations performed using Equations 1–3. Total simulation time = 24 hours. Parameters in Supplementary file 2. Model description and development in Methods. Inset: Simulations showing the final concentration of [vir] produced by P. aeruginosa as a function of initial density and at different absolute growth ratios. (B) Tipping point. Simulations showing the lowest initial fraction of P. aeruginosa (Cp) that results in a final density (Cp/Cs) of 1 after 24 hr as a function of the absolute growth ratio. In these simulations, metabolism (ε) for P. aeruginosa was varied but metabolism for S. aureus and maximum growth rates (μ) of both species were held constants. For simulations showing the effect of varying ε see Figure 4—figure supplement 1. (C) Simulations showing the temporal changes in the density of P. aeruginosa (Cp) and S. aureus (Cs). Top panels show the effect of increasing the relative initial density of Cp at the same absolute growth ratio. Bottom panels show the effect of increasing the absolute growth ratio while maintaining the same initial population densities (Cp = 0.5, Cs = 0.5). (D) Experimental data: The final density of a co-cultured population grown in TSB medium with different carbon sources plotted as a function of the initial percentage of P. aeruginosa. Standard error of the mean (SEM) from a minimum of three biological replicates. R2 and p values shown on plot are from a linear regression. * indicates a significant difference in the final density of P. aeruginosa and S. aureus (Mann-Whitney, p≤0.0463) (Shapiro-Wilk for normality ≤0.0001). When all final density ratios are compared within each carbon source p≤0.0029, Kruskal-Wallis. The exact number of biological replicates and all P values are in Supplementary file 5. Final cell density in CFU/mL shown in Figure 4—figure supplement 1. Data points outlined in black indicate the tipping point; the lowest initial percentage of P. aeruginosa examined that led to a significantly greater amount of P. aeruginosa after 24 hr of growth. Shaded region indicates 95% confidence interval. (E) Tipping point of the population defined as the first initial percentage of P. aeruginosa measured where there was not a significant difference in the final density of P. aeruginosa and S. aureus. Data from panel D. Number above each bar indicates absolute growth ratio calculated using data in Figure 1E. We did not find the tipping point for pyruvate and thus it is not included on this plot.
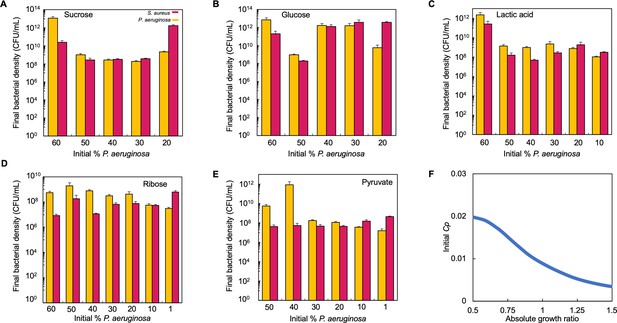
Raw bacteria density (CFU/mL) for co-cultures that were grown in TSB medium with different carbon sources.
Raw data for Figure 4D. (A–E) In each panel, the final density of P. aeruginosa and S. aureus in co-culture for 24 hr in the carbon source indicated is plotted. Standard error of the mean (SEM) from a minimum of three biological replicates. Exact number of biological replicates and all p values can be found in Supplementary file 5. (F) Tipping point. Simulations showing the lowest initial fraction of P. aeruginosa (Cp) that results in a final density ratio (Cp/Cs) of 1 after 24 hr as a function of the absolute growth ratio. In these simulations, metabolism (ε) for S. aureus was varied but metabolism for P. aeruginosa and maximum growth rates (μ) of both species were held constant.
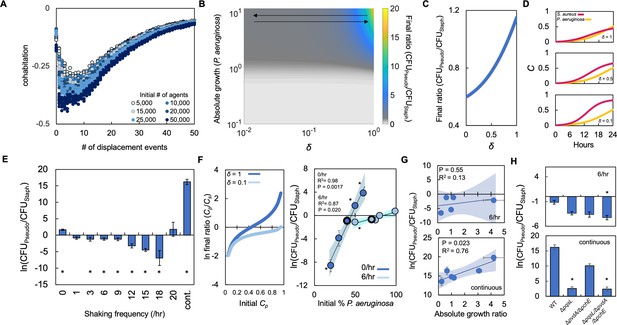
Periodically disturbing co-cultured populations of P. aeruginosa and S. aureus determines their co-existence.
(A) Simulation using our ABM showing the ‘cohabitation’ variable as a function of the number of disturbance events. These simulation trends were used to estimate trends in δ (Equations 1–3). Each simulation was performed 10 times; each data point represents an outcome from a single simulation. See Methods for ABM description and parameter justification. Sensitivity analysis showing U-shaped trend over a wide range of parameters displayed in Figure 5—figure supplement 1. (B) Heat map showing the effect of δ and absolute growth of P. aeruginosa (fixed absolute growth of S. aureus) on the final ratio of P. aeruginosa and S. aureus. For panels, B-D, simulations performed using Equations 1–3. Total simulation time = 24 hr. Parameters in Supplementary file 2. Model description and development in Methods. (C) Simulations showing the effect of changing δ on the final ratio of P. aeruginosa and S. aureus for a fixed ratio of absolute growth (1). (D) Simulations showing the temporal changes in the density of P. aeruginosa (Cp) and S. aureus (Cs) at different values of δ as indicated on each panel. (E) Experimental results. The ratio of P. aeruginosa to S. aureus as a function of disturbance events in TSB medium containing glucose as the carbon source. * indicates a significant difference (p≤0.0202 using a Mann-Whitney, Shapiro-Wilk<0.0001) in the final density of both populations. Kruskal-Wallis for all data points; p<0.0001. Raw density data (in CFU/mL) shown in Figure 5—figure supplement 2. Standard error of the mean (SEM) from a minimum of three biological replicates. For panels E, G, and H, the exact number of biological replicates and all P values are in Supplementary file 6. (F) Left: Simulations (Equations 1–3) showing the effect of changing δ on the final population density when the initial density of P. aeruginosa (Cp) is varied. Total simulation time = 24 hr. Parameters in Supplementary file 2. Right: Experimental data showing the final density of a co-cultured population either undisturbed (0 /hr, dark blue) or disturbed at 6 /hr (light blue) grown in TSB medium with different carbon sources plotted as a function of the initial percentage of P. aeruginosa. SEM from a minimum of three biological replicates. R2 and p values shown on plot are from a linear regression. * indicates a significant difference in the final density of P. aeruginosa and S. aureus (Mann-Whitney; p≤0.0202, Shapiro-Wilk; p=0.0190). Kruskal-Wallis for all data points: 0 /hr – p=0.0009; 6 /hr – p=0.115. Final cell density in CFU/mL shown in Figure 5—figure supplement 2. The exact number of biological replicates and all p values are in Supplementary file 6. Shaded regions indicate 95% confidence interval. (G) Experimental results. The ratio of P. aeruginosa to S. aureus after 24 hr of growth and with 6 disturbance events per hour (top) or in a continuously disturbed environment (bottom) in TSB medium containing different carbon sources. Difference between all data points: 6 /hr: p=0.0023 (Kruskal-Wallis); continuous: p<0.0366, (Kruskal-Wallis). Weighted least squares regression: 6 /hr, R2=0.034 p=0.765; continuous R2=0.72 p=0.0332. Raw cell density data (in CFU/mL) shown in Figure 5—figure supplement 2. SEM from a minimum of three biological replicates. Shaded regions indicate 95% confidence interval. (H) Experimental Results: The effect of six disturbance events per hour and continuous disturbance on the final density ratio of P. aeruginosa knockout strains and S. aureus wild type strain. Raw density data (in CFU/mL) shown in Figure 5—figure supplement 2. * indicates that the final density ratio was significantly different from co-culture with the wildtype strain (p<0.0129, Dunn test for joint ranks using wildtype as the control group). SEM from a minimum of five biological replicates.
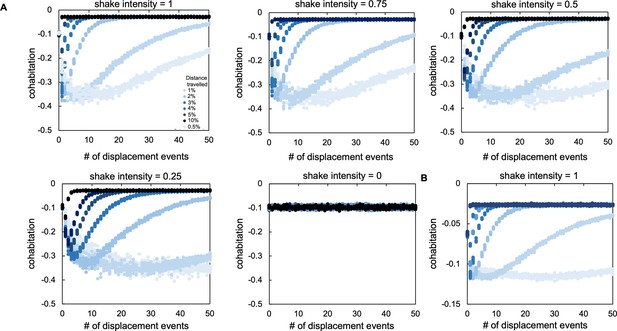
Sensitivity analysis from our agent-based model.
(A) The effect of changing the total percentage of agents that are moved in each displacement event (called shake intensity). A shake intensity value of 1 indicates that all agents were moved as a result of a shaking event. Each different colored series of circles indicates a different initial population density (as indicated in the legend). In panels A and B, for each set of parameters, simulations were performed 10 times; each data point represents the outcome of a single simulation. For panels A and B, ‘cohabitation’ is measured as the frequency of cohabited patches minus the expected frequency of cohabited patches (frequency of patches inhabited by P. aeruginosa agents x frequency of patches inhabited by S. aureus agents) such that a negative value means populations spatially overlap less than expected based on random chance. (B) The effect of adding dispersal through diffusion for P. aeruginosa agents in our ABM. P. aeruginosa can move 5% of the world (10 patches). This movement is separate from dispersal due to disturbance (shake intensity set at 1 for this plot). As in panel A, each different colored series of data points indicates the distance travelled by each agent after a disturbance event.

Raw bacteria density (CFU/mL) for co-cultures that were disturbed periodically.
(A) Final bacterial density for co-cultured populations grown in TSB with glucose. Standard error of the mean (SEM) from a minimum of three biological replicates. This is the raw data for Figure 5E. For panels A, D, E, F, G, H, and I, exact number of biological replicates and all P values can be found in Supplementary file 6. (B) Reducing the total initial density of bacteria in the population leads to qualitatively consistent findings. SEM from three biological replicates. p=0.78, two-tailed t-test, Shapiro-Wilk=0.74. (C) Raw density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in TSB medium containing various initial percentages of P. aeruginosa disturbed at 6 /hr. This raw data was used to produce Figure 5F, right panel. SEM from a minimum of three biological replicates. Exact number of biological replicates and all p values can be found in Supplementary file 6. (D) Raw density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in TSB medium containing various carbon sources disturbed at 6 /hr. SEM from a minimum of three biological replicates. (E) Raw density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in TSB medium containing various carbon sources that was continuously disturbed. SEM from a minimum of three biological replicates. (F) Final density ratio plotted as a function of the ratio of maximum growth rates (P. aeruginosa/S. aureus, left panel) or the ratio of [ATP] (P. aeruginosa/S. aureus, right panel) for populations disturbed at 6 /hr. Growth rates and [ATP] from Figure 1B and C, respectively. SEM from a minimum of three biological replicates. (G) Final density ratio plotted as a function of the ratio of maximum growth rates (P. aeruginosa/S. aureus, left panel) or the ratio of [ATP] (P. aeruginosa/S. aureus, right panel) for populations that were continuously disturbed. Growth rates and [ATP] from Figure 1B and C, respectively. SEM from a minimum of three biological replicates. (H) Raw density (CFU/mL) of S. aureus and knockout strains of P. aeruginosa in co-culture in TSB medium containing various carbon sources that were disturbed at 6 /hr. SEM from a minimum of four biological replicates. (I) Raw density (CFU/mL) of S. aureus and knockout strains of P. aeruginosa in co-culture in TSB medium containing various carbon sources that were continuously disturbed. SEM from a minimum of five biological replicates.
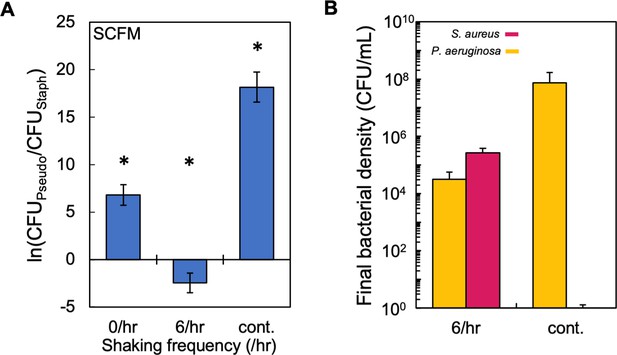
Periodic, but not continuous, disturbance can reduce dominance of P. aeruginosa in SCFM.
(I) Experimental results. The final density ratio of P. aeruginosa to S. aureus as a function of shaking frequency in SCFM containing glucose as the carbon source. * indicates a significant difference (p≤0.009 using a Mann-Whitney, Shapiro-Wilk <0.0001) in the final density of both populations. Kruskal-Wallis for all data points: p=0.0004 (Shapiro-Wilk; p<0.0001). Standard error of the mean (SEM) from a minimum of five biological replicates. Exact number of biological replicates and all p values can be found in Supplementary file 7. Note that if no colonies were observable after 72 hr on mannitol salt agar (selection for S. aureus), we assigned a value of 0.3 (translating to ~1 CFU/mL in 3 mL of medium), thus allowing us to ln transform the density for S. aureus. This adjustment is not reflected in panel B. (J) Raw density (CFU/mL) of S. aureus and P. aeruginosa in co-culture in SCFM containing glucose that was disturbed at 6 /hr or continuously. SEM from a minimum of five biological replicates. This data was used for panel A.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (S. aureus) | RN4220 | BEI resources | N/A | Wildtype strain |
| Strain (P. aeruginosa) | PA14 | Lingchong You | N/A | Wildtype strain |
| Strain (P. aeruginosa) | PA14 (ΔpqsL) | Geisel School of Medicine at Dartmouth (Hanover, NH); Orazi et al., 2019 | Knockout strain | |
| Strain (P. aeruginosa) | PA14 (ΔpvdA/ ΔpchE) | Geisel School of Medicine at Dartmouth (Hanover, NH); Orazi et al., 2019 | Knockout strain | |
| Strain (P. aeruginosa) | PA14 (ΔpqsL /ΔpvdA/ ΔpchE) | Geisel School of Medicine at Dartmouth (Hanover, NH); Orazi et al., 2019 | Knockout strain | |
| Commercial assay or kit (ATP assays) | BacTiter-Glo Microbial Cell Viability Assay | Promega | Cat. # G8230 | Used to measure ATP |
| Software, algorithm (MATLAB) | MATLAB with curve fitting and optimization toolboxes | MathWorks Inc. | N/A | Used to measure maximum growth rate |
| Software, algorithm (JMP Pro 16) | JMP Pro 16 | SAS Institute Inc. | N/A | Used for statistical analysis |
| Software, algorithm (Netlogo) | Netlogo | This paper; Tisue and Wilensky, 2004 | Used to measure spatial distribution | |
| Other (Aeraseal film) | Aeraseal sealing membranes | Sigma Aldrich | Cat #A9224 | Used to cover 6 well plates |
Additional files
-
Supplementary file 1
Statisical analysis for Figure 1.
(a) P values for data presented in Figure 1. n represents the number of biological replicates. To calculate absolute growth, all biological replicates from maximum growth rate and [ATP] were used. Thus, the value of n for absolute growth represents the smallest number of biological replicates included in the calculation.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp1-v3.docx
-
Supplementary file 2
Parameter set and statistical analysis for Figure 2.
(a) Base parameter set used in our mathematical model (Equations 1–3). (b) P values for data presented in Figure 2. n represents the number of biological replicates. Shapiro-Wilk for data sets in Figure 2A and B, P<0.0001. In Figure 2B and a Dunn test for joint rank was used to compare final density ratios. The final density ratio of the wildtype P. aeruginosa strain co-cultured with S. aureus was used as the control. TSB supplemented with glucose was used for these experiments.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp2-v3.docx
-
Supplementary file 3
Statisical analysis for Figure 2—figure supplement 1.
(a) P values for data presented in Figure 2—figure supplement 1, panel H. Shapiro-Wilk for final density ratios, P=0.0004; for final bacterial densities, P<0.0001. n represents the number of biological replicates.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp3-v3.docx
-
Supplementary file 4
Statistical analysis for Figure 3.
(a) P values for data presented in Figure 3. n represents the number of biological replicates. To calculate absolute growth, all biological replicates from maximum growth rate and ATP were used. Thus, the value of n for absolute growth represents the smallest number of biological replicates included in the calculation. Two-tailed t-tests were used for [ATP], growth rate and absolute growth measurements. A Mann-Whitney (Shapiro-Wilk, P<0.0001) was used to assess differences in the final density of P. aeruginosa and S. aureus (Figure 3E and J).
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp4-v3.docx
-
Supplementary file 5
P values for data presented in Figure 4.
Shapiro-Wilk, P≤0.0001 for all carbon sources. n represents the number of biological replicates.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp5-v3.docx
-
Supplementary file 6
Statistical analysis for Figure 5.
(a) P values for data presented in Figure 5E, G and F. Shapiro-Wilk for all data sets in this figure; P≤0.019. n represents the number of biological replicates. (b) P value for data presented in Figure 5F, right panel. Shapiro-Wilk for final density ratios, P<0.0001. Shapiro-Wilk for final bacterial densities, P=0.0190. n represents the number of biological replicates.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp6-v3.docx
-
Supplementary file 7
Statistical analysis for Figure 5—figure supplement 3.
(a) P values for data presented in Figure 5—figure supplement 3. Shapiro-Wilk for final density ratio and for bacterial densities, P<0.0001.
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp7-v3.docx
-
Supplementary file 8
Residual values for growth curve fitting.
(a) Average residual values for all curve fitting and biological replicates. Curve fitting was used to determine maximum growth rate in Figure 1B (TSB medium). (b) Average residual values for all curve fitting and biological replicates. Curve fitting was used to determine maximum growth rate in Figure 3A (AMM medium). (c) Average residual values for all curve fitting and biological replicates. Curve fitting was used to determine maximum growth rate in Figure 3F (SCFM medium).
- https://cdn.elifesciences.org/articles/83664/elife-83664-supp8-v3.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/83664/elife-83664-mdarchecklist1-v3.docx





