Structure of human phagocyte NADPH oxidase in the resting state
Figures
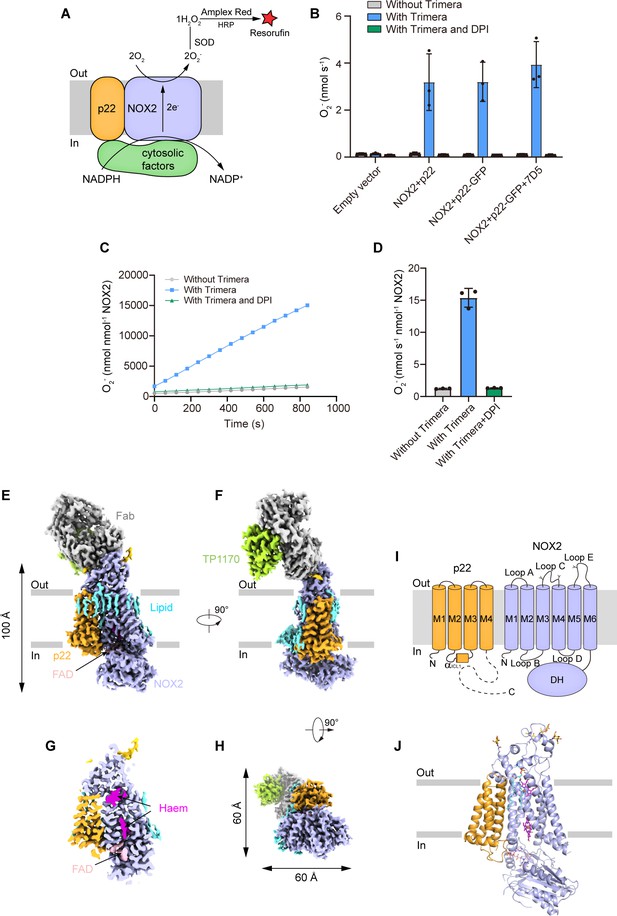
The structure of the human NOX2-p22 complex in the resting state.
(A) Schematic of the NOX2 enzymatic assay. O2- produced by NOX2 are converted into H2O2 by SOD. In the presence of H2O2, horseradish peroxidase (HRP) converts the nonfluorescent Amplex Red into resorufin, the fluorescence of which is measurable and proportional to the concentration of H2O2. (B) The activity of the NOX2-p22 complex in crude cell membrane measured using Amplex Red assay. The activity is determined by subtracting the background of the enzymatic assay buffer without crude cell membrane. Data are shown as means ± standard deviations; N=3 technical replicates. (C) The amounts of O2- produced by NOX2-p22-7D5-TP1170 complex in nanodisc in the presence of Trimera are plotted versus time. The represented data are shown. DPI, diphenyliodonium, NADPH oxidase inhibitor. (D) The rates of O2- production in C are summarized. Data are shown as means ± standard deviations; N=3 technical replicates. (E) Side view of the cryo-EM map of NOX2-p22-7D5-TP1170 complex. The approximate boundaries of the phospholipid bilayer are indicated as gray thick lines. p22 and NOX2 are colored in orange and light blue. 7D5 and TP1170 are colored in gray and light green. FAD, lipid, and glycosylation decoration are colored in pink, cyan, and gold, respectively. (F) A 90°-rotated side view compared to D. (G) The cut-open view of the cryo-EM map of NOX2-p22. Haem and FAD is colored in magenta and pink, respectively. (H) A 90°-rotated bottom view of E. (I) Topology of p22 and NOX2 subunits. Transmembrane helices are shown as cylinders, unmodeled disordered regions are shown as dashed lines. The phospholipid bilayer is shown as gray layers. Glycosylation sites were indicated by protrusions. DH, dehydrogenase domain of NOX2. (J) Structure of NOX2-p22 complex in cartoon representation. The colors are the same as in D.
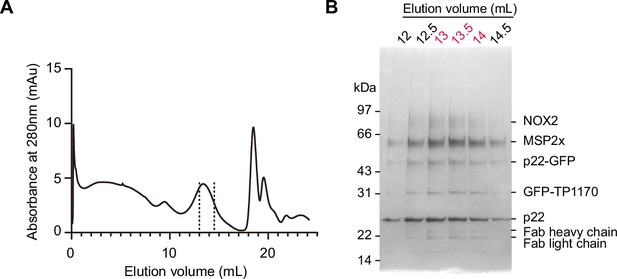
Protein purification.
(A) Size-exclusion chromatography profile of the NOX2-p22-7D5-TP1170 complex in nanodisc. Fractions between dashes were used for cryo-EM sample preparation. (B) Silver-stained SDS-PAGE gel of purified NOX2-p22-7D5-TP1170 protein in the nanodisc. Fractions with numbers in red were used for cryo-EM sample preparation. For uncropped gel, please see Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
The uncropped and unedited gels for Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/83743/elife-83743-fig1-figsupp1-data1-v2.zip
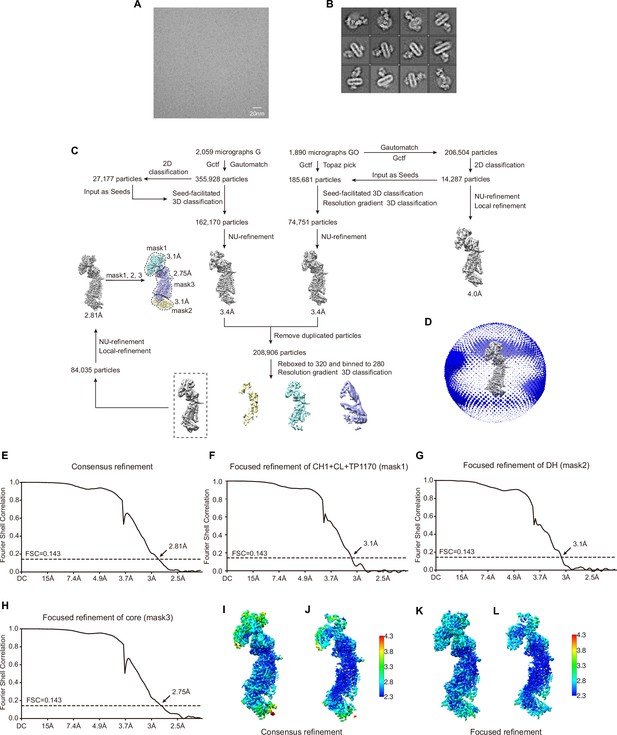
Image processing.
(A) Representative raw micrograph of NOX2-p22-7D5-TP1170 complex in nanodisc. (B) Two-dimensional (2D) class averages of NOX2-p22-7D5-TP1170 complex. (C) Workflow of the cryo-EM data processing. (D) Angular distributions of the final consensus reconstruction. (E) Gold-standard Fourier shell correlation curves of the final consensus refinement. (F) Gold-standard Fourier shell correlation curves of the focused refinement of the TP1170-constant region of Fab fragment (CH1 + CL + TP1170) using mask 1. (G) Gold-standard Fourier shell correlation curves of the focused refinement of the dehydrogenase (DH) domain using mask 2. (H) Gold-standard Fourier shell correlation curves of the focused refinement of the core domain using mask 3. (I) Local resolution of the NOX2-p22-7D5-TP1170 complex after consensus refinement. (J) Cut-open view of I. (K) Local resolution of the NOX2-p22-7D5-TP1170 complex after focused refinement. (L) Cut-open view of K.
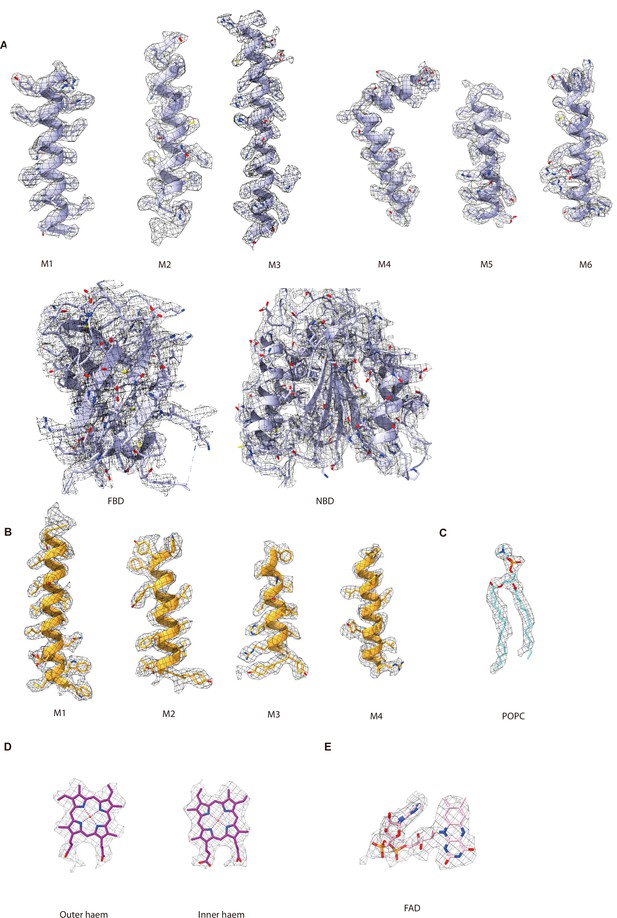
Representative local electron density maps contoured at 4.62σ.
(A) Local electron density maps of NOX2. (B) Local electron density maps of p22. (C) Local electron density of POPC. (D) Local electron density maps of haems bound in NOX2. (E) Local electron density map of FAD bound in FAD-binding sub-domain (FBD) of NOX2.
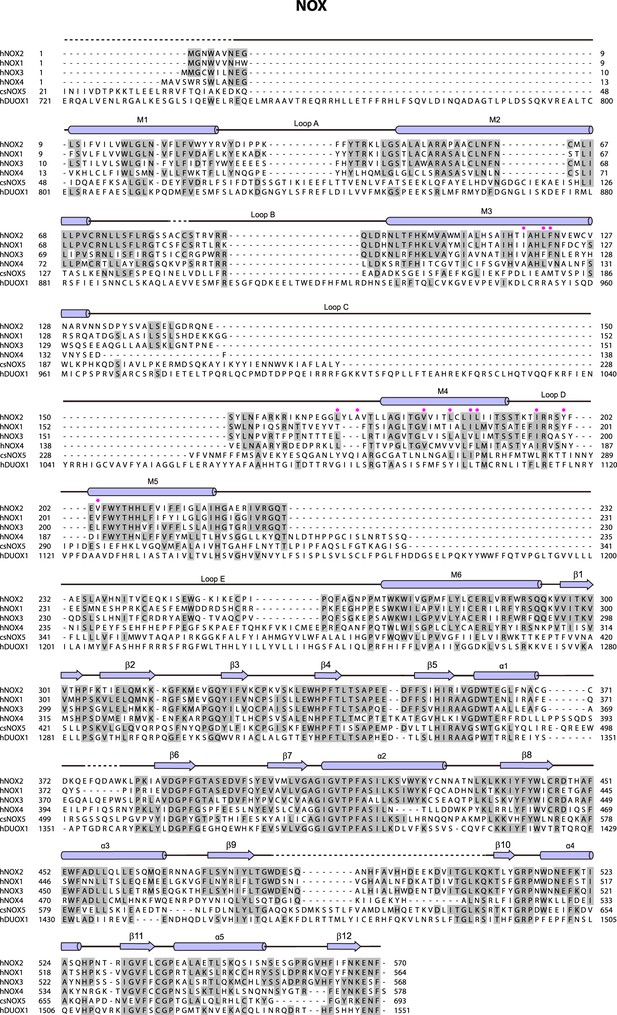
Sequence alignment of the NADPH oxidases (NOX) subunit.
The sequences of the Homo sapiens NOX2 (hNOX2, UniProtKB: P04839), H. sapiens NOX1 (hNOX1, UniProtKB: Q9Y5S8), H. sapiens NOX3 (hNOX3, UniProtKB: Q9HBY0), H. sapiens NOX4 (hNOX4, UniProtKB: Q9NPH5), Cylindrospermum stagnale NOX5 (csNOX5, UniProtKB: K9WT99) and H. sapiens DUOX1 (hDUOX1, UniProtKB: Q9NRD9) were aligned. The sequence alignment of Figure 1—figure supplements 4 and 5 are all shown as follows: Conserved residues are highlighted in gray. Secondary structures are indicated as cylinders (α helices), arrows (β sheets), and lines (loops). Unmodeled residues are indicated as dashed lines. The color of arrows and cylinders is the same as in Figure 1I. Residues interact with p22 are indicated as pink-filled circles.
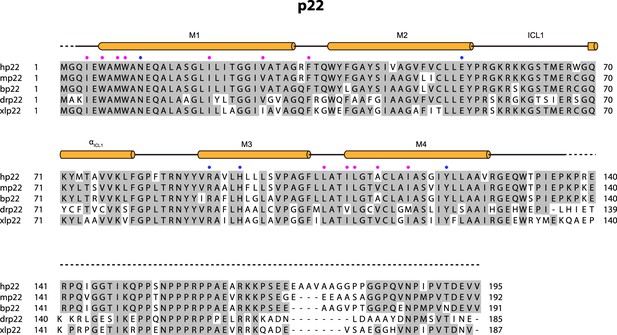
Sequence alignment of p22 subunit.
The sequences of the Homo sapiens p22 (hp22, UniProtKB: P13498), Mus musculus p22 (mp22, UniProtKB: Q61462), Bos taurus p22 (bp22, UniProtKB: O46521), Danio rerio p22 (drp22, UniProtKB: Q6PH62), and Xenopus laevis p22 (xlp22, UniProtKB: Q6AZJ1) were aligned. Residues interacting with NOX2 are indicated as pink-filled circles. The blue-filled circles indicate the residues that form a polar interaction network.
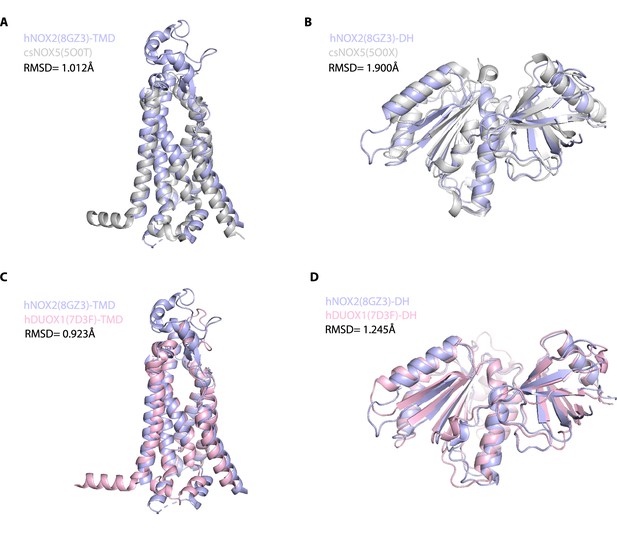
Structural alignment of NADPH oxidases (NOX) family proteins.
(A) Structural comparison of the transmembrane domain (TMD) of hNOX2 (light blue) and csNOX5 (gray). (B) Structural comparison of the dehydrogenase (DH) domain of hNOX2 (light blue) and csNOX5 (gray). (C) Structural comparison of the TMD of hNOX2 (light blue) and hDUOX1 (pink). (D) Structural comparison of the DH domain of hNOX2 (light blue) and hDUOX1 (pink).
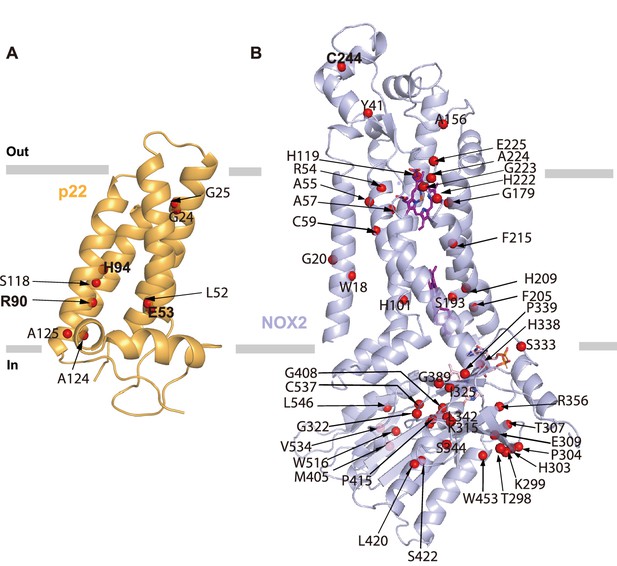
Locations of chronic granulomatous disease (CGD) mutations found in human patients.
(A) Mutations of NOX2 found in CGD patients (annotated in UNIPROT) are mapped onto the structural model of NOX2. Cα positions of mutations are shown as red spheres, NOX2 is shown in a light blue cartoon. (B) Mutations of p22 found in CGD patients (annotated in UNIPROT) are mapped onto the structural model of p22. Cα positions of mutations are shown as red spheres. p22 is shown in a bright orange cartoon. Mutations mentioned in the text are highlighted in bold.
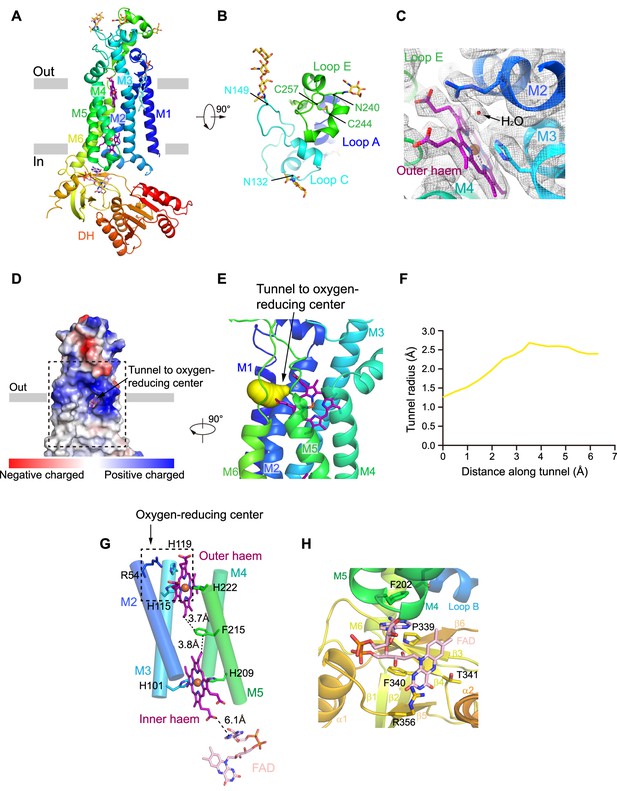
The structure of human NOX2.
(A) Structure of NOX2 in cartoon representation, and is colored in the rainbow pattern (N-terminus is blue and C-terminus is red). The phospholipid bilayer is shown as gray layers. (B) A 90°-rotated top view of A. The disulfide bonds and glycosylation are shown as sticks. Three extracellular loops are indicated. (C) Electron density at the oxygen-reducing center contoured at 6.3σ. The density of water is indicated with an arrow. The color is the same as in A. (D) Surface electrostatic potential distribution of NOX2 estimated by pymol. The tunnel to the oxygen reducing center is indicated with an arrow. Haem and glycosylation are shown as sticks. The phospholipid bilayer is shown as gray layers. (E) Side view of the oxygen-reducing center as shown in a dashed box in D. The predicted tunnel for the entry of oxygen substrate and release of superoxide anions is colored in yellow and indicated with an arrow. (F) Calculated tunnel radius shown in E. The starting point for calculation is the oxygen-reducing center. (G) The components along the electron transfer pathway of NOX. The edge-to-edge distances between adjacent components are shown in dashes. The oxygen reducing center is indicated as a dashed rectangle. The ligands are shown as sticks. The ferric ions are shown as spheres. The helices are shown as cylinders. The colors are the same as in A. (H) The close-up view of the FAD-binding site. Side chains of residues interacting with FAD are shown as sticks.
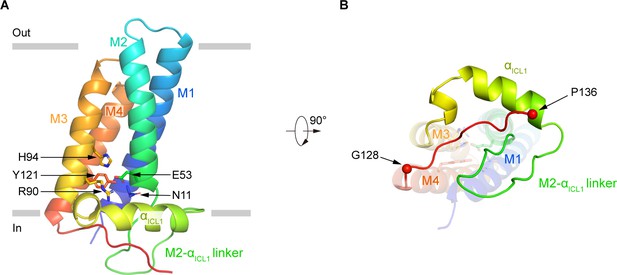
The structure of human p22.
(A) Structure of p22 in cartoon representation, and is colored in the rainbow pattern (N-terminus is blue and C-terminus is red). The phospholipid bilayer is shown as gray layers. (B) The 90°-rotated bottom view of A. The 128–136 segment is colored in red. Cα positions of G128 and P136 are indicated as spheres.
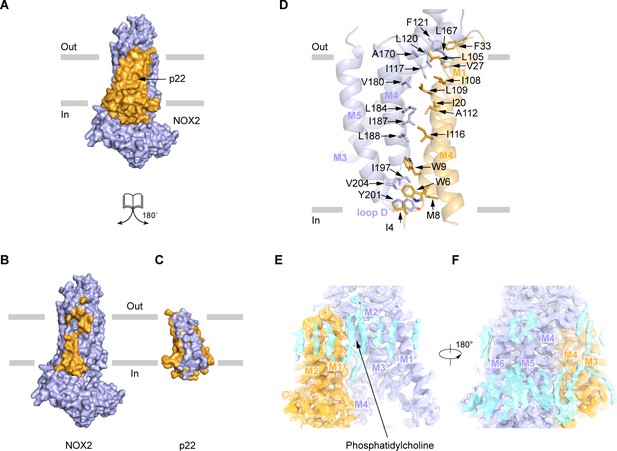
The interaction between NOX2 and p22.
(A–C) The open-book view of the interface between NOX2 and p22 in surface representation. NOX2 and p22 are colored in light blue and orange respectively in (A). Residues on NOX2 interacting with p22 are colored in orange in (B). Residues on p22 interacting with NOX2 are colored in light blue in (C). The phospholipid bilayer is shown as gray layers. (D) The interface between NOX2 and p22 in cartoon representation. Residues participating in the interaction between NOX2 and p22 are shown as sticks. (E) Putative lipids that are close to NOX2-p22 interface. The map is contoured at 5.5 σ. (F) A 180°-rotated view of E.
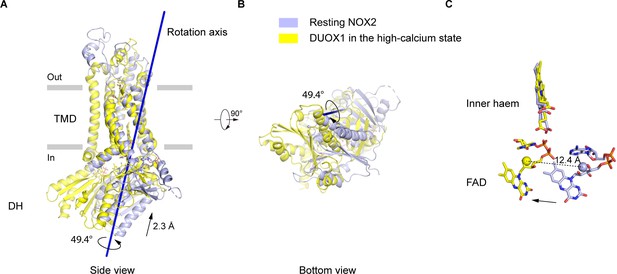
The movements of the dehydrogenase (DH) domain from the resting NOX2 to the active DUOX1.
(A) Side view of the structural alignment of NOX2 in the resting state (light blue) and DUOX1 in the high-calcium state (yellow) in cartoon representation. Their transmembrane domains (TMD) were used in alignment. The rotation axis of DH domains is shown in dark blue. The rotational and translational movements from the DH domain of NOX2 to the DH domain of DUOX1 are indicated with arrows. (B) Bottom view of A. (C) The centers of mass of FAD molecules in the resting NOX2 and in the active DUOX1 are shown as spheres and the movement of FAD is indicated by an arrow.
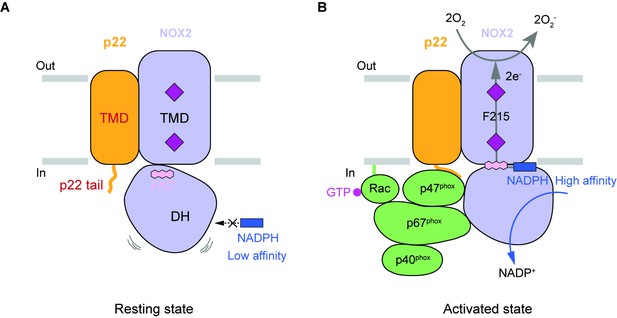
Hypothetic model for NOX2 activation.
(A) Resting state of the phagocyte NADPH oxidase. NOX2, p22, and cytosolic factors are shown as cartoon and colored the same as Figure 1A. (B) Activated state of the phagocyte NADPH oxidase. The electron transfer pathway is indicted with gray arrows.
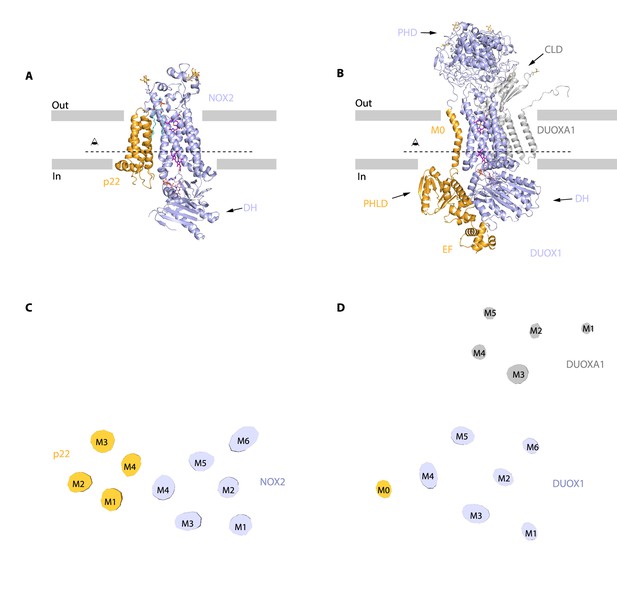
Distinct interaction mode between NOX2-p22 complex and DUOX1-DUOXA1 complex.
(A) Structure of human NOX2 and p22 complex (PDB ID: 8GZ3) in cartoon representation. The colors of each individual domain are the same as in Figure 1I. The approximate boundaries of the phospholipid bilayer are indicated as gray thick lines. Sugar moieties, haems, FAD, and lipids are shown as gold, purple, pink, and aquamarine sticks, respectively. (B) Structure of human DUOX1-DUOXA1 complex (PDB ID: 7D3F) in cartoon representation. PHD: peroxidase homology domain of DUOX1; PHLD: pleckstrin homology-like domain of DUOX1; EF: EF-hand calcium-binding module of DUOX1; DH: dehydrogenase domain of DUOX1; CLD: claudin-like domain of DUOXA1. The M0 helix, PHLD, and EF are colored in orange, and the other part of DUOX1 is colored in blue. The DUOXA1 is colored in gray. (C) Top view of the cross-section of NOX2-p22 cryo-EM map on the transmembrane layer at the position indicated as a dashed line in A. For clarity, the cryo-EM map was low-pass filtered to 7 Å. (D) Top view of the cross-section of DUOX1-DUOXA1 cryo-EM map on the transmembrane layer at the position indicated as a dashed line in B.
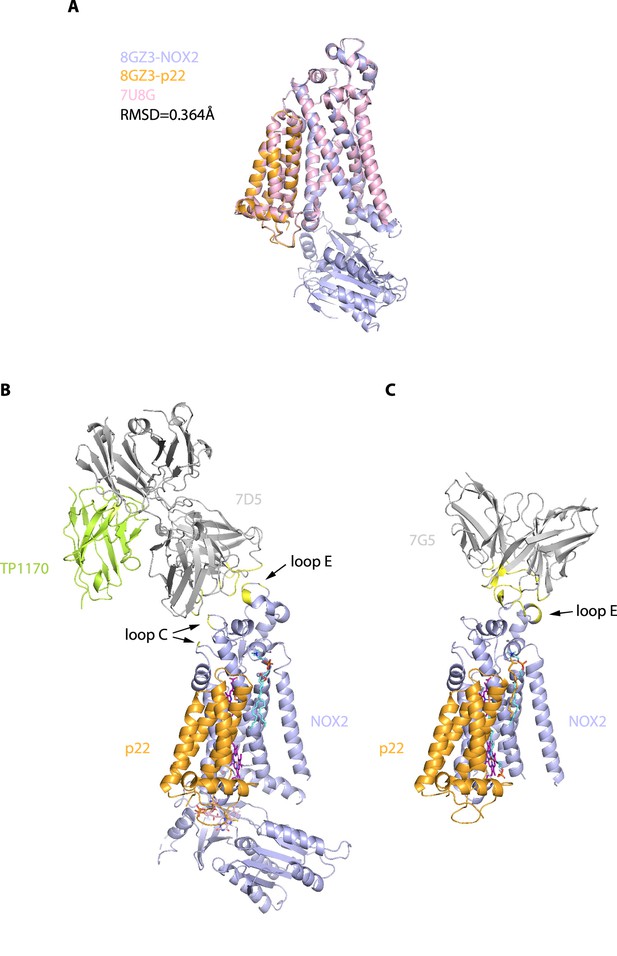
Structural comparison between NOX2-7D5 complex and NOX2-7G5 complex.
(A) Overall structural comparison of human NOX2-7D5 complex between this study (orange and blue, PDB ID: 8GZ3) and NOX2-7G5 complex (pink, PDB ID: 7U8G). (B) Structure of NOX2-7D5 complex in cartoon representation. The colors of each individual domain are the same as in Figure 1I. The residues participate the interaction between NOX2 and 7D5 are colored in yellow. (C) Structure of NOX2-7G5 complex colored the same as in B (PDB ID: 7U8G).
Tables
Cryo-EM data collection, refinement, and validation statistics.
| PDB IDEMDB ID | NOX2-p22-7D5-TP11708GZ3EMD-34389EMD-34390*/EMD-34620†/EMD-34622‡/EMD-34621§ |
|---|---|
| Data collection and processing | |
| Magnification | ×165,000 |
| Voltage (kV) | 300 |
| Electron exposure (e-/Å2) | 37.6 |
| Defocus range (μm) | –1.5 to –2.0 |
| Pixel size (Å) | 0.821 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 541,609 |
| Final particle images (no.) | 84,035 |
| Map resolution (Å) | 2.81*/3.1†/3.1‡/2.75§ |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 250–2.8 |
| Refinement | |
| Initial model used (PDB code) | Alpha Fold 2 |
| Model resolution (Å) | 2.8 |
| FSC threshold | 0.143 |
| Model resolution range (Å) | 250–2.8 |
| Map sharpening B factor (Å2) | –89.6*/–128.9†/–123.3‡/100.1§ |
| Model composition | |
| Non-hydrogen atoms | 8,542 |
| Protein residues | 1,214 |
| Ligands | 9 |
| B factors (Å2) | |
| Protein | 55.46 |
| Ligand | 58.94 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.531 |
| Validation | |
| MolProbity score | 1.60 |
| Clashscore | 5.77 |
| Poor rotamers(%) | 1.74 |
| Ramachandran plot | |
| Favored (%) | 97.49 |
| Allowed (%) | 2.42 |
| Disallowed (%) | 0.08 |
-
*
The values of consensus refinement.
-
†
The values of focused refinement of mask1 (CH1 + CL + TP1170).
-
‡
The values of focused refinement of mask2 (DH domain).
-
§
The values of focused refinement of mask3 (core).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CYBA | GenBank | AB590173.1 | |
| Gene (Homo sapiens) | CYBB | GenBank | NM_000397.4 | |
| Gene (Homo sapiens) | p47phox | GenBank | NM_000265.7 | |
| Gene (Homo sapiens) | p67phox | GenBank | LT740922.1 | |
| Gene (Homo sapiens) | rac1 | GenBank | NM_006908.5 | |
| Cell line (Spodoptera frugiperda) | Sf9 | Thermo Fisher Scientific | Cat. # 12659017 | RRID:CVCL_0549 |
| Cell line (Homo sapiens) | FreeStyle 293F | Thermo Fisher Scientific | Cat. # R79007 | RRID:CVCL_D603 |
| Transfected construct (human) | NOX2-p22 complex | This paper | Transfected construct (human) | |
| Antibody | Anti-NOX2 7D5 (Mouse monoclonal) | MBL Life Science | Cat. # D162-3 | |
| Recombinant DNA reagent | p22-GFP (plasmid) | This paper | GFP version of p22phox | |
| Recombinant DNA reagent | GFP-TP1170 (plasmid) | This paper; Pleiner et al., 2018 | PMID:29263082 | GFP version of TP1170 |
| Peptide, recombinant protein | SOD | Sigma-Aldrich | Cat. # S2515 | |
| Commercial assay or kit | In-Fusion HD Cloning | Takara Bio | Cat. # 639650 | |
| Chemical compound, drug | Amplex Red | GeneCopoeia | Cat. # C291 | |
| Chemical compound, drug | NADPH | Cayman Chemical | Item No. 9000743 | |
| Software, algorithm | SWISS-MODEL | SWISS-MODEL | RRID:SCR_013032 | |
| Software, algorithm | Chimera | Chimera | RRID:SCR_004097 | |
| Software, algorithm | Coot | Coot | RRID:SCR_014222 | |
| Software, algorithm | Phenix | Phenix | RRID:SCR_014224 |





