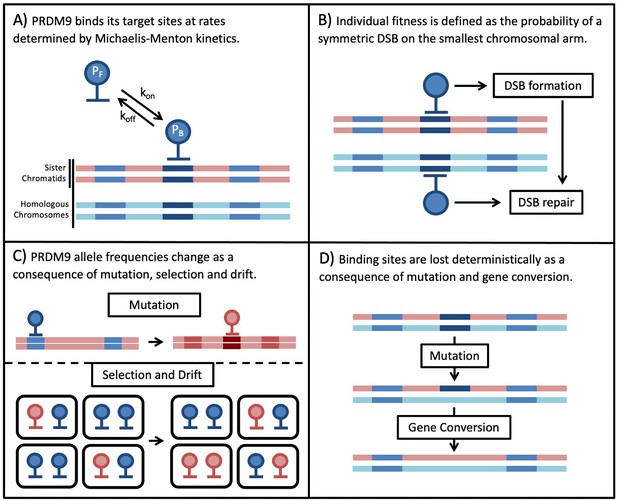
Down the Penrose stairs, or how selection for fewer recombination hotspots maintains their existence
Figures
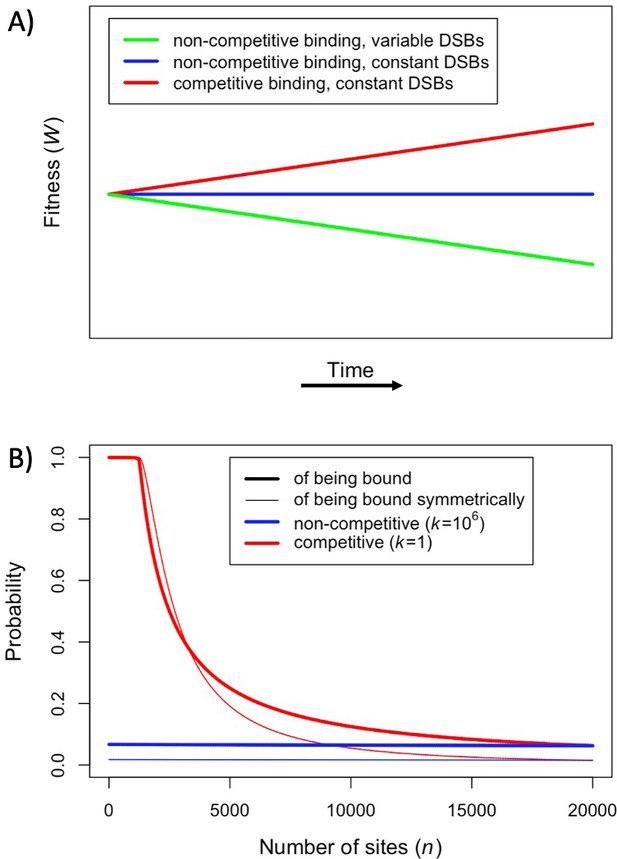
Fitness in the model with one heat.
(A) Cartoon depicting the qualitative change in fitness as a function of time under different models, assuming that all individuals are homozygous for a single PRDM9 allele (without turnover) and that all binding sites have the same binding affinity. For visualization purposes, we present results for a linear function. (B) The probability of an individual binding site being bound (thicker lines) or of a given locus being bound symmetrically (thinner lines) if all sites are very weak and non-competitive (; blue) or very strong and competitive (; red). The values shown were computed using Equation 1, Equation S2.2.2 where, for sake of comparison, in both cases we set the number of PRDM9 molecules such that 5000 sites would be bound in the presence of 20,000 binding sites (roughly consistent with observations in mice Parvanov et al., 2017; Grey et al., 2017). When binding sites are very weak, most PRDM9 molecules are not bound and therefore our model behaves as if there were no competition between binding sites.
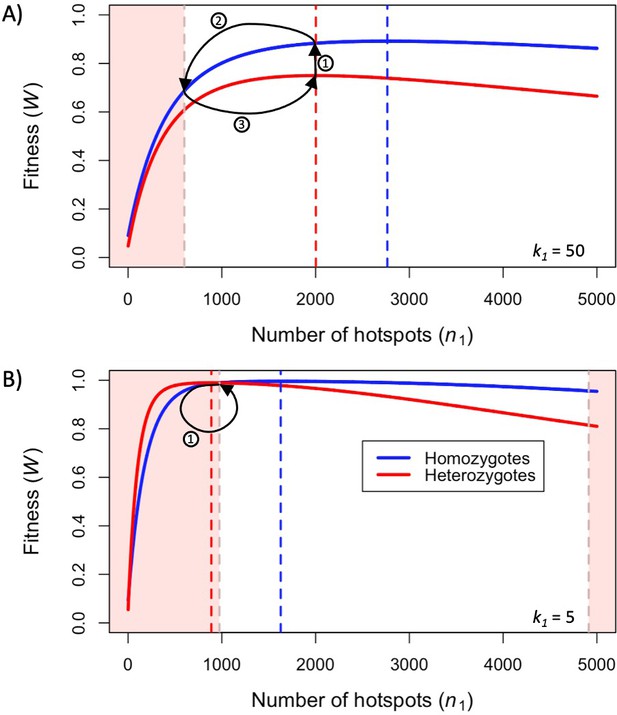
Fitness in the model with two heats.
Fitness as a function of the number of hotspots, for an individual homozygous for a PRDM9 allele (blue), or heterozygous for two PRDM9 alleles with the same number of hotspots (red), when considering (A) weak hotspots or (B) strong ones. Fitness was calculated using Equations 5 and 6, assuming a backdrop of weak binding sites that would bind 99% of PRDM9 molecules in the absence of hotspots. Vertical dashed lines indicate the number of hotspots that maximize the fitness of individuals homozygous (blue) or heterozygous (red) for the allele. The area shaded in light red indicates the number of hotspots where the fitness of an individual homozygous for PRDM9 has lower fitness than the maximal fitness of a heterozygous individual that carries one copy of the same PRDM9 allele (where the other allele has the optimal number of hotspots for heterozygotes). In both panels, the region shaded in light red on the left reflects the case wherein the homozygous individual will have reduced fitness as a consequence of PRDM9 binding to weak binding sites. In panel B, the region on the right reflects the case wherein the homozygous individual will have reduced fitness as a consequence of limited symmetric binding at hotspots. The numbered arrows illustrate the behavior of the simplified evolutionary dynamic described in the text.
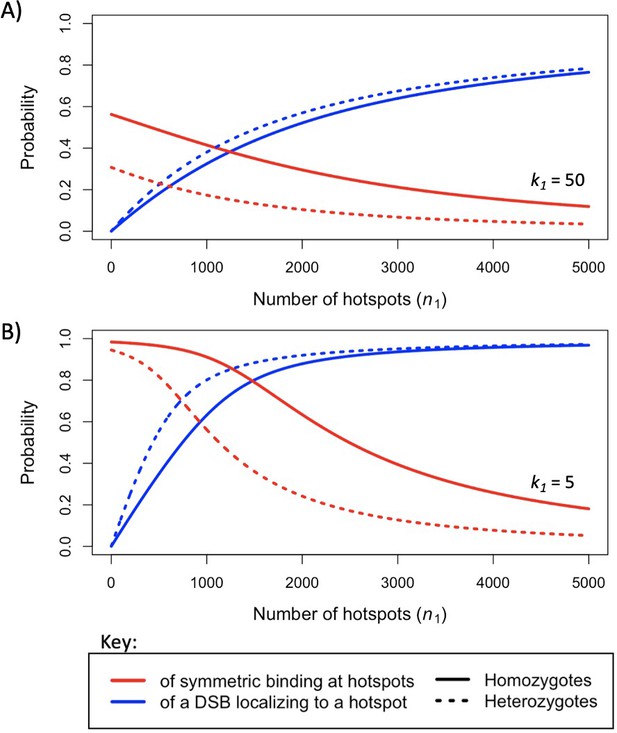
The probability that a hotspot has been symmetrically bound (red), and the probability (or proportion) of DSBs localizing to hotspots (blue), when considering hotspots with (A) a relatively strong dissociation constant, or (B) a weaker one, under the two-heat model in individuals homozygous (solid lines) or heterozygous (dashed lines) for PRDM9.
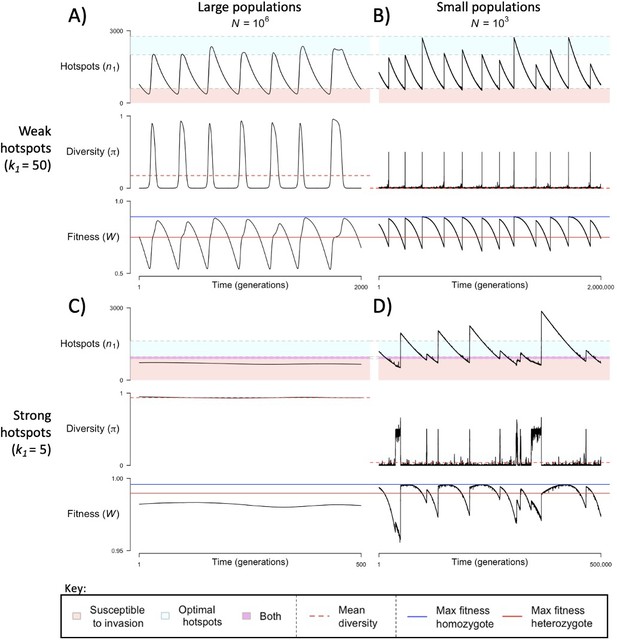
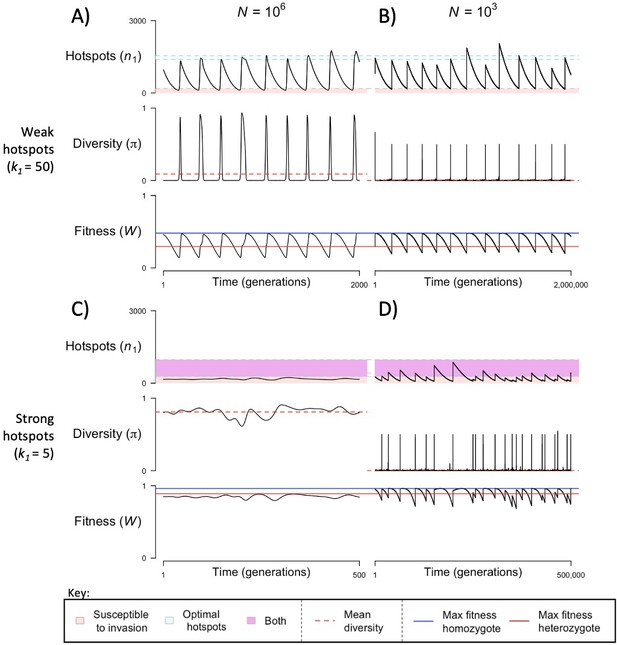
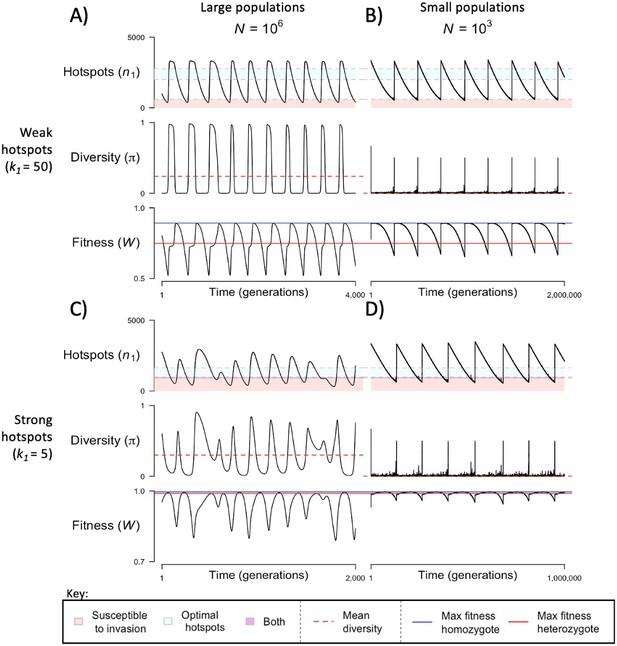
The dynamics of the model with two heats.
The mean number of hotspots (), diversity at PRDM9 (), and mean fitness (), as a function of time. We show four cases: with weak and strong hotspots (top and bottom rows, respectively) and large and small population size (left and right column, respectively). Simulations (as detailed in the model section) were run with the specified population sizes and hotspot dissociation constants, with other parameter values detailed in Table 1. The time (x-axis) in each plot has been scaled by the duration of PRDM9 turnover to allow for ~10 ‘cycles’ (see Figure 3), which span ~103 times more generations in small populations compared to large ones. The highlighted range of hotspot numbers in which a PRDM9 allele is ‘susceptible to invasion’ corresponds to the regions shaded in red in Figure 3. In turn, the highlighted range of hotspot numbers designated as ‘optimal hotspots’ corresponds to the range between the number that optimizes fitness in heterozygotes and the number that optimizes fitness in homozygotes (between the dashed lines shown in Figure 3). See Appendix 4 for analogous results assuming an alternative value for the number of PRDM9 molecules expressed during meiosis (), and Appendix 5 for analogous results assuming a tight unimodal distribution of the number of hotspots associated with newly arising PRDM9 alleles. This figure was generated using Figure 4—source data 1–4.
-
Figure 4—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 55,000 simulated generations of the two-heat model with a large population size () and weak hotspots ().
This data was used to generate Figure 4 and Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig4-data1-v2.zip
-
Figure 4—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots ().
This data was used to generate Figure 4 and Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig4-data2-v2.zip
-
Figure 4—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 55,000 simulated generations of the two-heat model with a large population size () and strong hotspots ().
This data was used to generate Figure 4 and Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig4-data3-v2.zip
-
Figure 4—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500,000 simulated generations of the two-heat model with a small population size () and strong hotspots ().
This data was used to generate Figure 4 and Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig4-data4-v2.zip
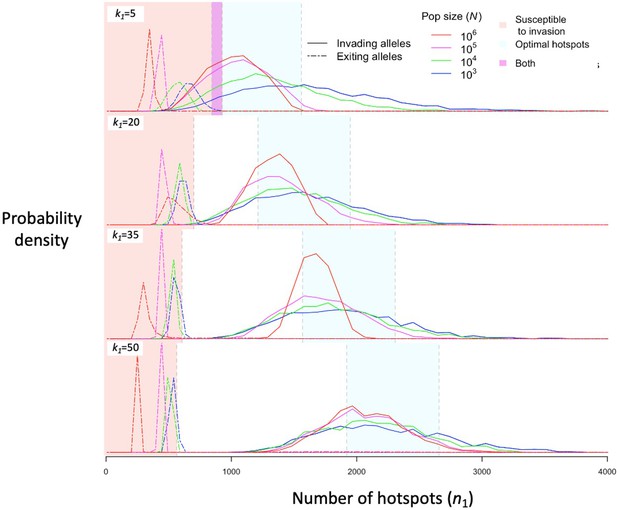
The distribution of initial and final numbers of hotspots among segregating PRDM9 alleles.
The relative densities of initial and final numbers of hotspots (corresponding to invading and exiting alleles, respectively) for different dissociation constants (rows) and population sizes (line colors) obtained from simulations of five million PRDM9 allele trajectories with each set of parameters (see the model section for details about the simulations and Table 1 for other parameter values). Shaded regions are defined the same way as in Figure 4. This figure was generated using Figure 5—source data 1–2.
-
Figure 5—source data 1
The distribution of the initial number of hotspots () among PRDM9 alleles, weighted by their mean allele frequency and sojourn time.
The first two columns represent population size () and the dissociation constant at hotspots () respectively. The remaining 50 columns describe the distribution in bins, with each bin corresponding to a range of 100 initial hotspots (e.g., the first column represents the probability of drawing an allele from the population at random which initially began with between 1 and 100 hotspots). This data was used to generate Figure 5.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig5-data1-v2.zip
-
Figure 5—source data 2
The distribution of the number of hotspots () among exiting PRDM9 alleles, weighted by their mean allele frequency and sojourn time.
The first two columns represent population size () and the dissociation constant at hotspots () respectively. The remaining 100 columns describe the distribution in bins, with each bin corresponding to a range of 50 initial hotspots (e.g., the first column represents the probability of drawing an allele from the population at random which would ultimately leave the population with between 1 and 50 hotspots). This data was used to generate Figure 5.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig5-data2-v2.zip
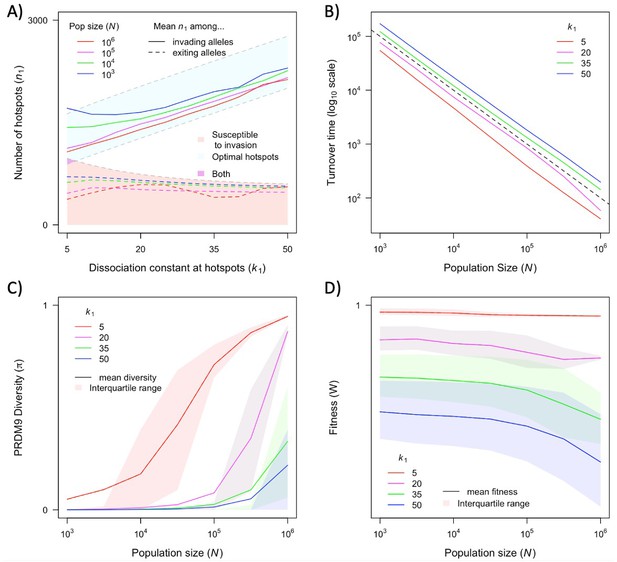
The dependence of key quantities on population size and hotspot heat.
(A) Mean number of hotspots among incoming and exiting alleles (corresponding to the distributions shown in Figure 5). (B) Average turnover time of segregating PRDM9 alleles (see text). (C) Mean and interquartile range of diversity at PRDM9 over time. (D) Mean and interquartile range of fitness in the population. Each point in these graphs is based on simulations of 5x106 PRDM9 alleles (see model section for details) with the specified parameter values and the other model parameter values specified in Table 1. See Appendix 4 for analogous results, assuming alternative levels of PRDM9 expression. This figure was generated using Figure 6—source data 1 and 2.
-
Figure 6—source data 1
Summary statistics from simulations for a range of population sizes () and hotspot dissociation constants (), including mean turnover times, mean initial numbers of hotspots, and mean number of hotspots for exiting PRDM9 alleles (each weighted by the sojourn time of PRDM9 alleles).
This data was used to generate Figure 6.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-data1-v2.zip
-
Figure 6—source data 2
Additional summary statistics from simulations for a range of population sizes () and hotspot dissociation constants (), including mean diversity at PRDM9 () and population fitness (), and the inter-quartile ranges of each.
This data was used to generate Figure 6.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-data2-v2.zip
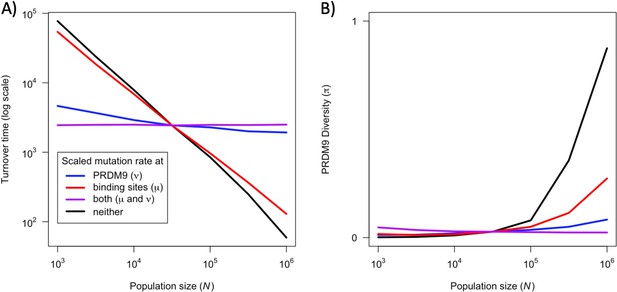
The effect of population size on the rate of turnover (A) and diversity (B) at PRDM9 while keeping population-scaled mutation rates at PRDM9, binding sites, or both, constant (when ).
This figure was generated using Figure 6—figure supplement 1—source data 1–2.
-
Figure 6—figure supplement 1—source data 1
Mean PRDM9 turnover times for a range of population sizes ( to 106) when considering moderately strong hotspots (), under normal parameters, when the population-scaled mutation rate at PRDM9 has been normalized to that of a moderate population size (), when the population-scaled mutation rate at PRDM9 binding sites has been normalized to that of a moderate population size (), and when both mutation rates have been so normalized.
This data was used to generate Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp1-data1-v2.zip
-
Figure 6—figure supplement 1—source data 2
Mean values of diversity at the PRDM9 locus () for a range of population sizes ( to 106) when considering moderately strong hotspots (), under normal parameters, when the population-scaled mutation rate at PRDM9 has been normalized to that of a moderate population size (), when the population-scaled mutation rate at PRDM9 binding sites has been normalized to that of a moderate population size (), and when both mutation rates have been so normalized.
This data was used to generate Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp1-data2-v2.zip
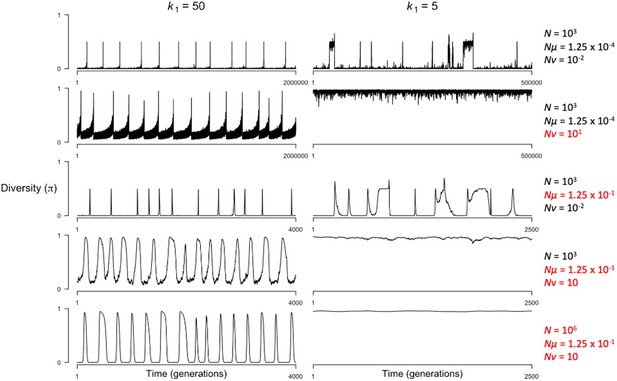
The effect of population-scaled mutation rates on the dynamics of the model when hotspots are cold () or hot ().
Values for population size () and mutation rates at PRDM9 binding sites () and at the PRDM9 locus () used in each simulation are shown to the right of each row. Values shown in black indicate those typical for small populations () and values shown in red indicate those typical for large populations (). This figure was generated using Figure 4—source data 1–4 and Figure 6—figure supplement 2—source data 1–6.
-
Figure 6—figure supplement 2—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the population-scaled mutation rate at PRDM9 was adjusted to match that of a large population (with ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data1-v2.zip
-
Figure 6—figure supplement 2—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the population-scaled mutation rate at PRDM9 was adjusted to match that of a large population (with ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data2-v2.zip
-
Figure 6—figure supplement 2—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the population-scaled mutation rate at PRDM9 binding sites was adjusted to match that of a large population (with ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data3-v2.zip
-
Figure 6—figure supplement 2—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the population-scaled mutation rate at PRDM9 binding sites was adjusted to match that of a large population (with ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data4-v2.zip
-
Figure 6—figure supplement 2—source data 5
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 55,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the population-scaled mutation rate at both PRDM9 and PRDM9 binding sites was adjusted to match that of a large population (with ; ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data5-v2.zip
-
Figure 6—figure supplement 2—source data 6
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 55,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the population-scaled mutation rate at both PRDM9 and PRDM9 binding sites was adjusted to match that of a large population (with ; ; ).
This data was used to generate Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-fig6-figsupp2-data6-v2.zip
Dynamics of the two-heat model when .
The mean number of hotspots (), diversity at PRDM9 (), and mean fitness (), as a function of time. We show four cases: with weak and strong hotspots (top and bottom rows respectively) and large and small population size (left and right column respectively). See Figure 4 and Appendix 4 for details. This figure was generated using Appendix 4—figure 1—source data 1–4.
-
Appendix 4—figure 1—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 2,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data1-v2.zip
-
Appendix 4—figure 1—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data2-v2.zip
-
Appendix 4—figure 1—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500 simulated generations of the two-heat model with a large population size () and strong hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data3-v2.zip
-
Appendix 4—figure 1—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data4-v2.zip
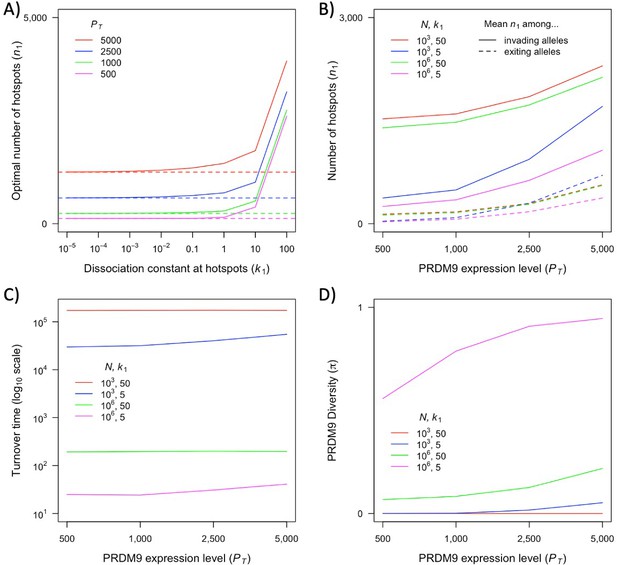
The dependence of key quantities on PRDM9 expression level.
(A) The number of hotspots which optimizes fitness for an individual homozygous for PRDM9 alleles as a function of the dissociation constant at hotspots () for different levels of PRDM9 expression (). Horizontal dashed lines indicate the expected optimal values as approaches zero (). (B) Mean number of hotspots among incoming and exiting alleles in small or large population sizes (=103 or 106) and when hotspots are relatively strong or weak ( = 5 or 50), as a function of PRDM9 expression level (). (C) Average turnover time of segregating PRDM9 alleles for different parameters as a function of PRDM9 expression level (). (D) Mean diversity at PRDM9 over time for different parameters as a function of PRDM9 expression level (). The weighting of PRDM9 alleles in B and C is as detailed in the main text. This figure was generated using Appendix 4—figure 2—source data 1.
-
Appendix 4—figure 2—source data 1
Summary statistics from simulations for large and small population sizes ( or 106) and for small and large hotspot dissociation constants ( or 50), for a range of values of the expression level of PRDM9 ( or 5000), including mean turnover times, mean initial numbers of hotspots, and mean number of hotspots for exiting PRDM9 alleles (each weighted by the sojourn time of PRDM9 alleles), as well as the mean and interquartile range of diversity at PRDM9 (). This data was used to generate Appendix 4—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig2-data1-v2.zip
Dynamics of the two-heat model when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
The mean number of hotspots (), diversity at PRDM9 (), and mean fitness (), as a function of time. We show four cases: with weak and strong hotspots (top and bottom rows respectively) and large and small population size (left and right column respectively). See Figure 4 and Appendix 5 for details. This figure was generated using Appendix 5—figure 1—source data 1–4.
-
Appendix 5—figure 1—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 4,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data1-v2.zip
-
Appendix 5—figure 1—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data2-v2.zip
-
Appendix 5—figure 1—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000 simulated generations of the two-heat model with a large population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data3-v2.zip
-
Appendix 5—figure 1—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 1,000,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data4-v2.zip
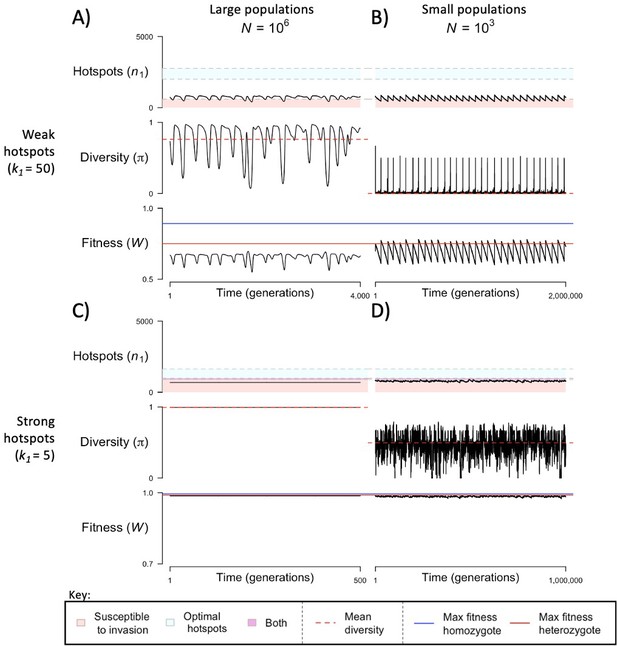
Dynamics of the two-heat model when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
The mean number of hotspots (), diversity at PRDM9 (), and mean fitness (), as a function of time. We show four cases: with weak and strong hotspots (top and bottom rows respectively) and large and small population size (left and right column respectively). See Figure 4 and Appendix 5 for details. This figure was generated using Appendix 5—figure 2—source data 1–4.
-
Appendix 5—figure 2—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 4,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data1-v2.zip
-
Appendix 5—figure 2—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data2-v2.zip
-
Appendix 5—figure 2—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500 simulated generations of the two-heat model with a large population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data3-v2.zip
-
Appendix 5—figure 2—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 1,000,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data4-v2.zip
Tables
Parameters and variables of the model.
| Parameter | Description | Value |
|---|---|---|
| Diploid population size | 103–106 | |
| μ | Mutation rate from hot to cold alleles at PRDM9 binding sites per generation | 1.25x10–7 |
| Mutation rate at the PRDM9 locus per generation | 10–5 | |
| Number of DSBs initiated per meiosis | 300 | |
| Total number of PRDM9 molecules expressed per meiosis | 5,000 | |
| Dissociation constant of PRDM9 binding sites with affinity | 5–50 | |
| Probability that a conversion tract spans the PRDM9 binding motif | 0.7 | |
| Proportion of the genome corresponding to the smallest chromosome | 1/40 | |
| Variable | Description | |
| Number of PRDM9 binding sites with affinity , recognized by PRDM9 allele , across the genome | ||
| Probability that a given site with binding affinity , recognized by PRDM9 allele , is bound by PRDM9 in an individual with PRDM9 alleles and (Equation 1) | ||
| Total number of bound cognate PRDM9 molecules in an individual with PRDM9 alleles and | ||
| Number of free cognate PRDM9 molecules in an individual with PRDM9 alleles and per meiosis (Equation 2) | ||
| Probability that any PRDM9-bound site experiences a DSB in an individual with PRDM9 alleles and (Equation 3) | ||
| Probability of a PRDM9 binding site with affinity , recognized by PRDM9 allele , being symmetrically bound by PRDM9 and experiencing a DSB in an individual with PRDM9 alleles and (Equation 4) | ||
| The fitness of an individual with PRDM9 alleles and (Equation 5 for homozygotes, Equation 6 for heterozygotes) | ||
| The marginal fitness of PRDM9 allele across possible genotypes (Equation 7) | ||
| The allele frequency of PRDM9 allele (Equation 8) | ||
| Probability or rate of a PRDM9 binding site with affinity , recognized by PRDM9 allele , experiencing gene conversion spanning the PRDM9 binding motif, in an individual with PRDM9 alleles and (Equation 9) | ||
| Probability or rate of a PRDM9 binding site with affinity , recognized by PRDM9 allele , experiencing gene conversion spanning the PRDM9 binding motif, across the population (Equation 10) | ||
Additional files
-
Appendix 4—figure 1—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 2,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data1-v2.zip
-
Appendix 4—figure 1—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data2-v2.zip
-
Appendix 4—figure 1—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500 simulated generations of the two-heat model with a large population size () and strong hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data3-v2.zip
-
Appendix 4—figure 1—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), and a smaller number of expressed PRDM9 molecules ().
This data was used to generate Appendix 4—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig1-data4-v2.zip
-
Appendix 4—figure 2—source data 1
Summary statistics from simulations for large and small population sizes ( or 106) and for small and large hotspot dissociation constants ( or 50), for a range of values of the expression level of PRDM9 ( or 5000), including mean turnover times, mean initial numbers of hotspots, and mean number of hotspots for exiting PRDM9 alleles (each weighted by the sojourn time of PRDM9 alleles), as well as the mean and interquartile range of diversity at PRDM9 (). This data was used to generate Appendix 4—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app4-fig2-data1-v2.zip
-
Appendix 5—figure 1—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 4,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data1-v2.zip
-
Appendix 5—figure 1—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data2-v2.zip
-
Appendix 5—figure 1—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000 simulated generations of the two-heat model with a large population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data3-v2.zip
-
Appendix 5—figure 1—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 1,000,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 10 non-degenerate sites.
This data was used to generate Appendix 5—figure 1.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig1-data4-v2.zip
-
Appendix 5—figure 2—source data 1
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus (pi) per generation, across 4,000 simulated generations of the two-heat model with a large population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data1-v2.zip
-
Appendix 5—figure 2—source data 2
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 2,000,000 simulated generations of the two-heat model with a small population size () and weak hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data2-v2.zip
-
Appendix 5—figure 2—source data 3
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 500 simulated generations of the two-heat model with a large population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data3-v2.zip
-
Appendix 5—figure 2—source data 4
The number of hotspots (), average population fitness (), and diversity at the PRDM9 locus () per generation, across 1,000,000 simulated generations of the two-heat model with a small population size () and strong hotspots (), when the distribution of the initial numbers arises from a motif with 11 non-degenerate sites.
This data was used to generate Appendix 5—figure 2.
- https://cdn.elifesciences.org/articles/83769/elife-83769-app5-fig2-data4-v2.zip