Gating interactions steer loop conformational changes in the active site of the L1 metallo-β-lactamase
Figures
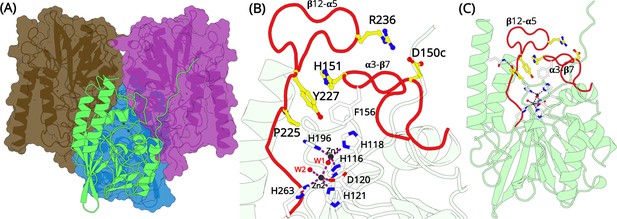
Overall structural features of the L1 metallo-β-lactamase (MBL).
(A) The homo-tetrameric conformation of L1 MBL (PDB entry 1SML). Three monomers are illustrated in surface representation; the fourth monomer is shown in green cartoon. (B) A close up of the zinc-binding site and the elongated α3-β7 and β12-α5 loops (red). Two zinc ions are present at the binding site. Zn1 coordinates with residues H116, H118, H196, and a water molecule forming a tetrahedral geometry. Zn2 is trigonal bipyramidal and coordinates with residues D120, H121, and H263. Zn1 and Zn2 are bridged by a water molecule. The position of D150c, H151, P225, Y227, and R236 are shown as yellow sticks. (C) L1 monomer illustrated as cartoon (green); the position of the two elongated loops (red) that forms a part of the active site in each monomer are highlighted in red.
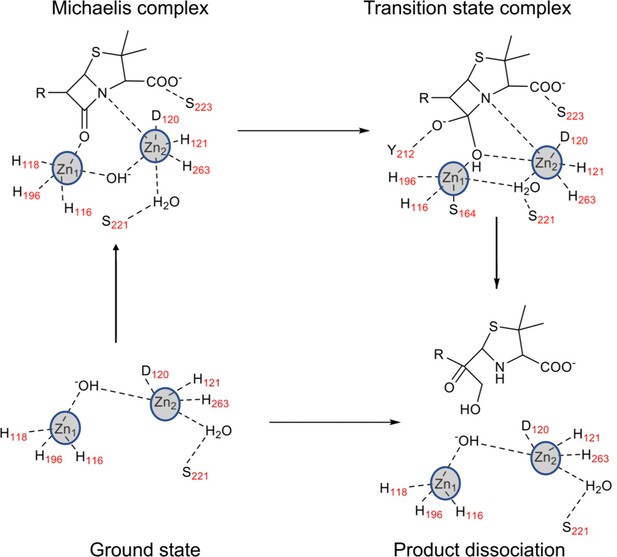
Catalytic cycle of L1 metallo-β-lactamase (MBL).
Proposed mechanism of β-lactam hydrolysis by S. maltophilia L1 MBL.
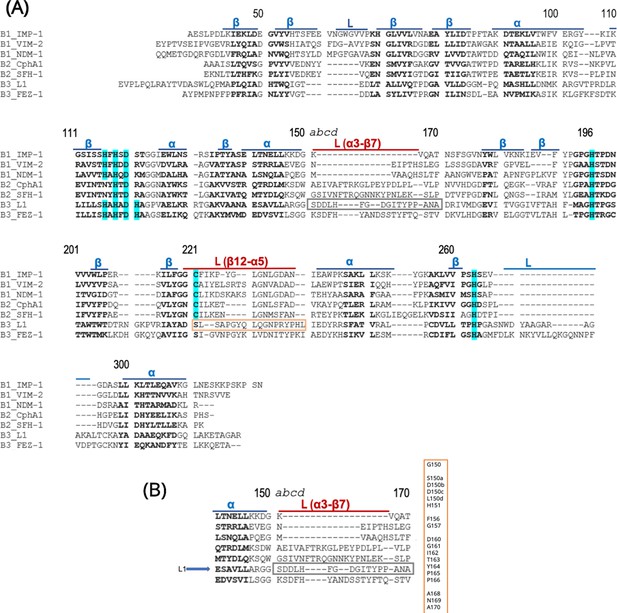
Sequence alignment of seven class B metallo-β-lactamase with the BBL numbering.
(A) Conserved secondary structure elements for the three subclasses are indicated above the sequences: α-helix, β-sheets, and L-loop. Amino acid insertions in the newly sequences enzymes are represented by lowercase letters. The zinc ligands in at least one subclass are shaded in cyan. L(α3-β7) and L(β12-5) sequences are highlighted in red represent largely variable regions. The 14 sequence fragments off structurally conserved position, which cover the entirety of all sequences, are shown in boldface. (B) The variable numbering around L(β12-α5). Figure 1—figure supplement 2 is adapted from Figure 1 of Garau et al., 2004.
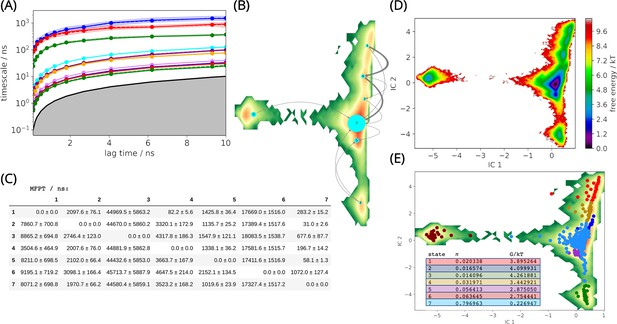
L1 metallo-β-lactamase Markov State Model.
(A) Implied timescales (ITS) plot. The ITS plot describes the convergence behavior of the implied timescales associated with the processes at the lag time of 5 ns. (B) Net flux pathways plot highlights the transition pathways in the relevant directions, between the highest energy state 1, and all other states. The thickness of the arrows between states suggests the possibility of each transition. The thicker the arrow, the greater the possibility. (C) Mean first passage times between metastable states. (D) Free energy landscape. The sampled free energy projected onto the first two time-lagged independent components (ICs) at lag time τ = 5.0 ns. (E) The distribution of cluster centers highlighting the presence of the metastable states. Macrostate distributions of conformations projected onto the first two ICs identified seven macrostates. The population of each state (π) and its free energy estimation is listed.
-
Figure 2—source data 1
Mean first passage time.
- https://cdn.elifesciences.org/articles/83928/elife-83928-fig2-data1-v1.xlsx
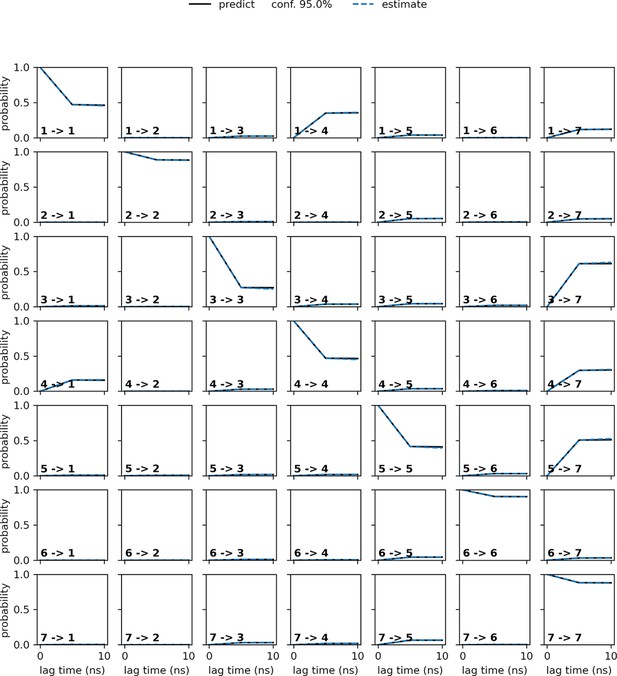
Chapman–Kolmogorov (CK) test plot.
The CK test plot, along with the ITS plot (Figure 2A) was used to assess the Markovian behavior of the constructed model.
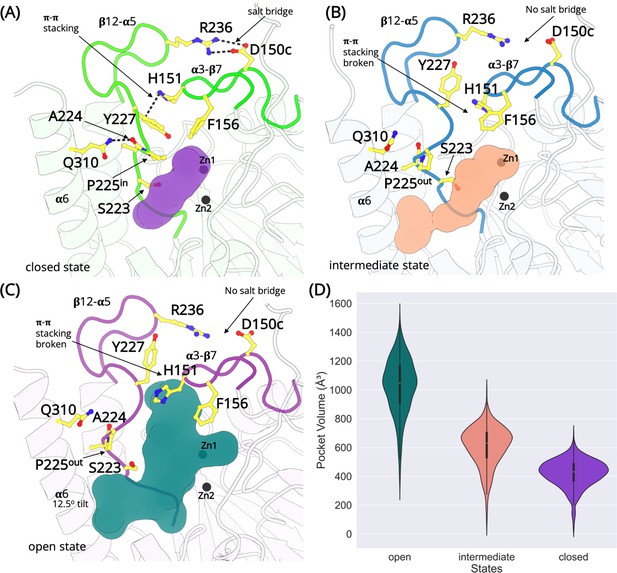
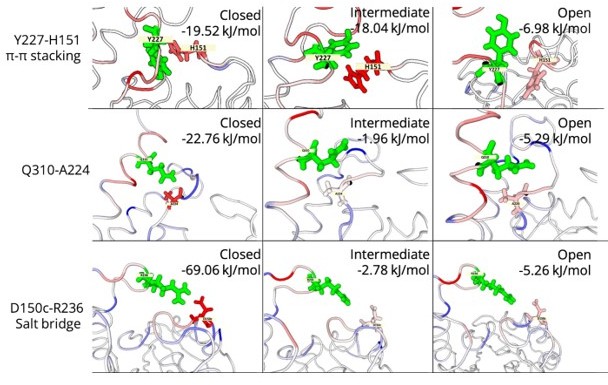
The L1 metallo-β-lactamase active site remodeling.
Structures exist in the closed, intermediate, and open states defined by a salt bridge between D150c-R236, π–π stacking interaction between H151-Y227 and the conformation of the cyclic side chain of P225. (A) In the closed state, the salt bridge and the π–π stacking interaction is formed and P225 adopts the ‘in’ conformation. The β12-α5 loop is collapsed occluding the active site (purple). (B) In the intermediate open state, the interactions are lost and P225 adopts the ‘out’ conformation. However, the β12-α5 loop is not completely in the open conformation. A small cavity between α6 helix and β12-α5 is formed that merges with the active site (pink). (C) In the fully open state, the salt bridge and the π–π stacking interaction is lost. P225 adopt ‘the out’ conformation. The β12-α5 loop moves outwards, opening a large cavity that merges with the active site (teal). (D) The calculated volume of the active site in different states.
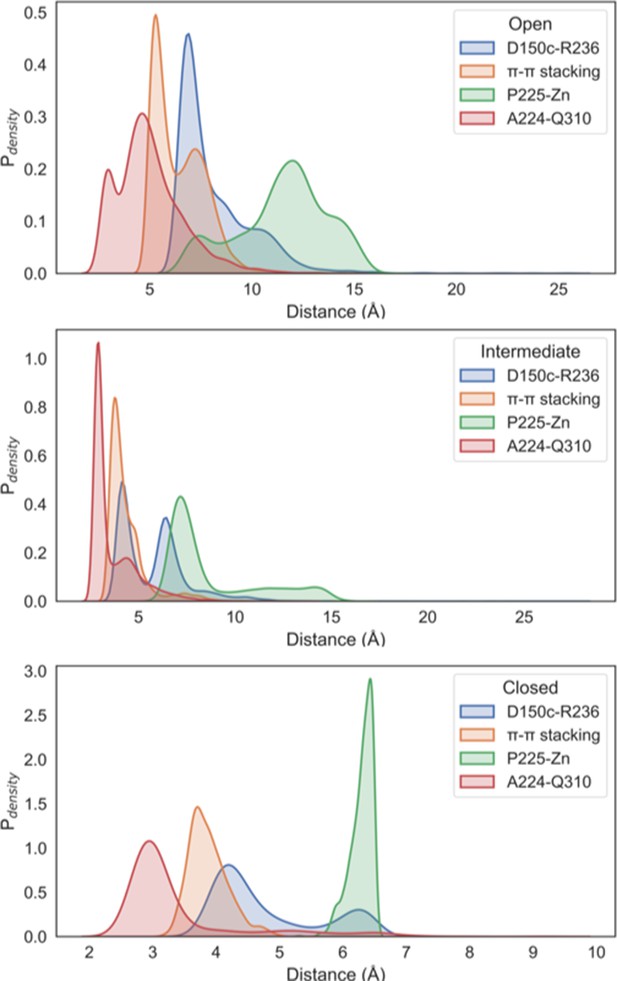
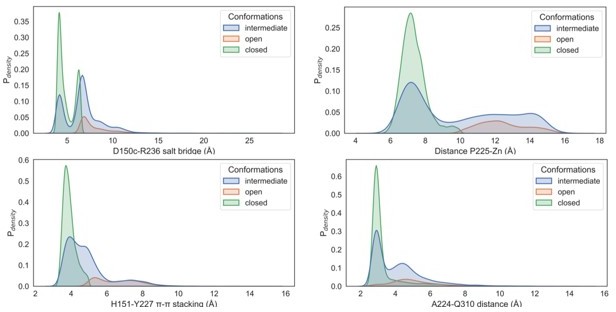
Probability weighted distance distributions in the open, intermediate, and the closed states.
The distances between the D150c-R236 salt bridge (blue), H151-Y227 π–π stacking (orange), A224-Q310 (red), and P225-Zn (green) have been illustrated.
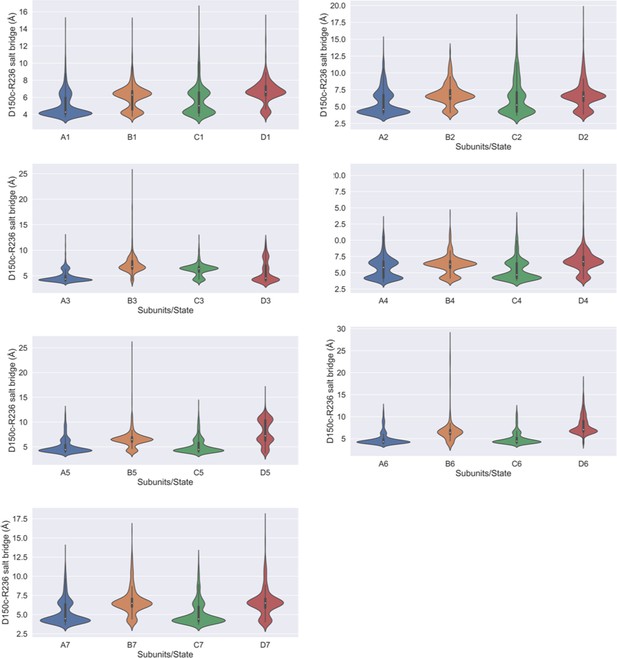
Salt bridge interaction between D150c-R236.
Violin plots highlighting the calculated D150c-R236 salt bridge interaction in each of the four subunits in different metastable states.
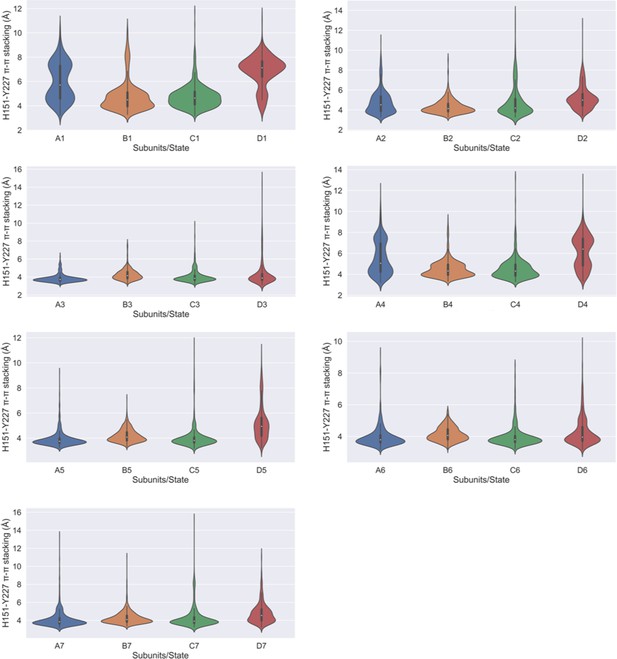
π–π stacking interaction between H151-Y227.
Violin plots highlighting the calculated H151-Y227 π–π stacking interaction in each of the four subunits in different metastable states.
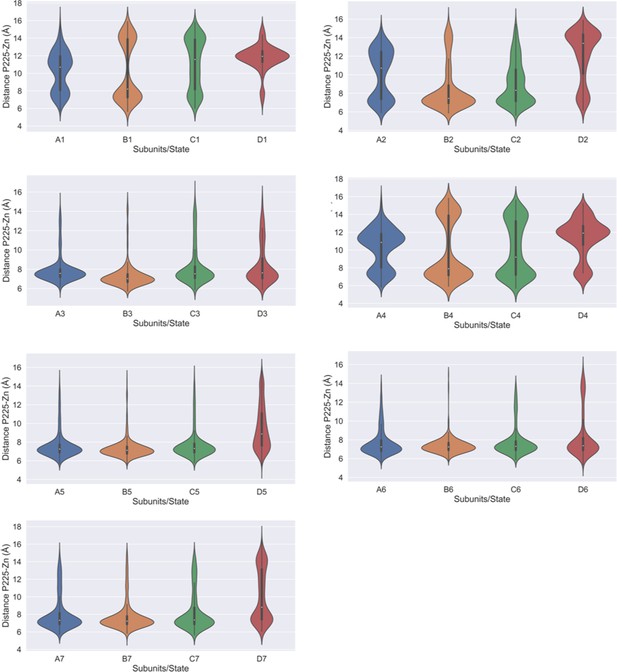
Distance between P225 and the centroid of zinc ions.
Violin plots highlighting the calculated P225-Zn distance in each of the four subunits in different metastable states.
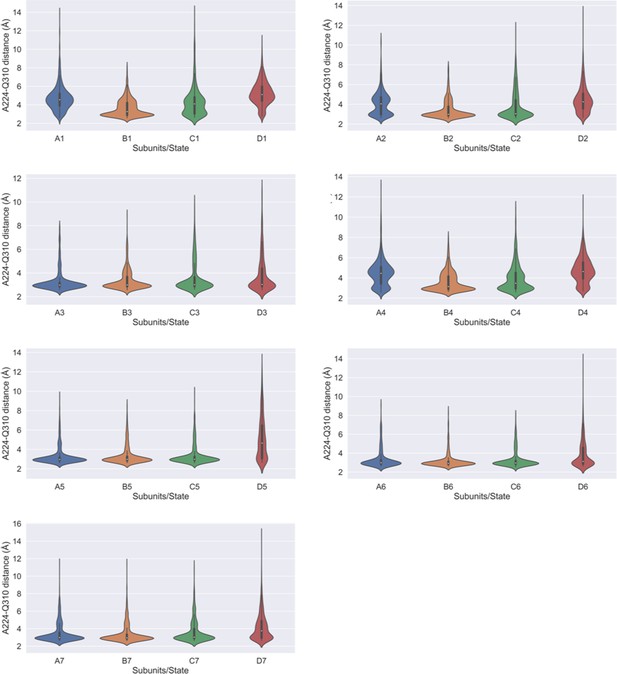
Interaction between A224-Q310.
Violin plots highlighting the calculated A224(bb)-Q310(sc) distance in each of the four subunits in different metastable states.

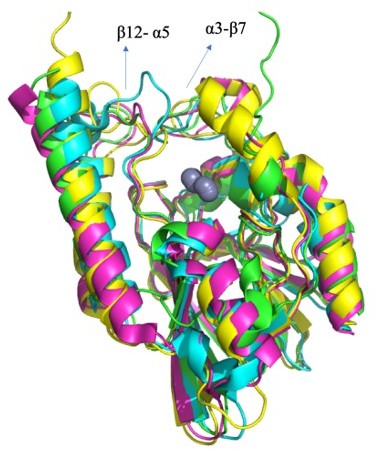
Structural comparison of the L1 metallo-β-lactamases.
A Cα-RMSD superimposition of state 7 (green) with a monomer from apo state crystal structure (PDB id 1SML; orange; 0.79 Å) and with inhibitor complex (PDB id 5DPX; purple; 0.77 Å) structure of L1 β-lactamase. The black balls represent the position of zinc ions.
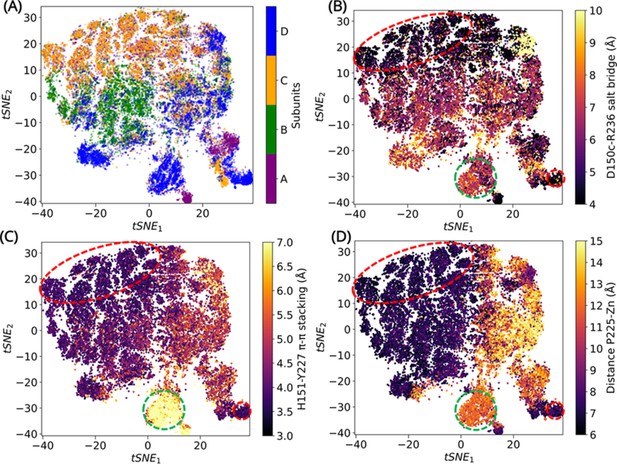
Convolutional variational autoencoder (CVAE)-based deep learning analysis.
CVAE-learned features of high-dimensional data represented in 2D following t-distributed stochastic neighbor embedding (tSNE) treatment. (A) The results show that A and C subunits show similar dynamics, while B and D subunits cluster together. The (B) salt bridge interactions between D150c-R236, (C) H151-Y227 π–π stacking interaction, and (D) distance between P225 and the centroid of Zn atoms have been plotted. The red box highlights a region where closed confromations are found, while the green box highlights a region of open conformation. The other conformations are open intermediate (two or more features are lost) or closed intermediate (two or more features are present).
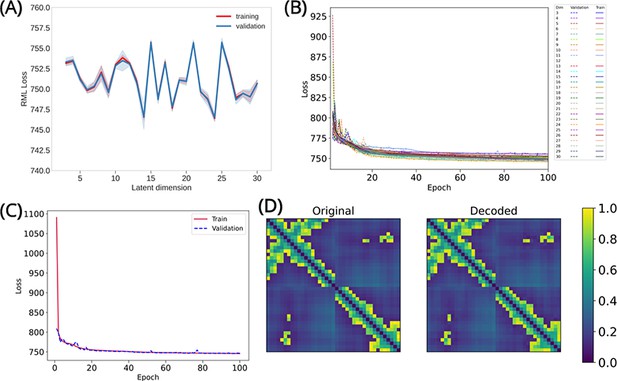
Convolutional variational autoencoder (CVAE)-based deep learning analysis.
(A) The training and validation loss plotted as assessed simultaneously over consecutive epochs at various reduced dimensions. Dimension 14 was selected for further analysis as it displayed the lowest loss. (B) The validation loss during CVAE implementation is plotted at different latent dimension for determining optimum values of the low dimension. The loss was calculated over 100 epochs from dimensions 3 to 30. (C) The convergence of training and validation loss from dimension 14, as assessed over 100 epochs. (D) Comparison between original input data and reconstructed data (decoded) are illustrated to ensure no loss of essential information during the compression and reconstruction process.
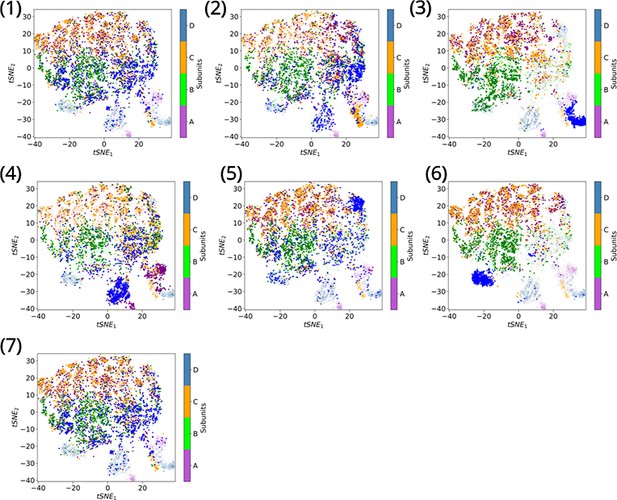
Clustering metastable states via t-distributed stochastic neighbor embedding (tSNE).
Individual subunits are plotted on the tSNE space after clustering of states 1–7. A comparison with Figure 4 highlights the open (state 1), intermediate open (state 4), intermediate closed (state 2), and closed conformations (state 7). Subunits A and C display similar dynamics while subunits B and D cluster together. The open state is best represented by subunit D of state 1, where D150c-R236 salt bridge is broken, H151-Y227 π–π stacking is lost and P225 adopts an out conformation. The closed state is best represented by subunit C in state 7. Subunit A in state 4 represents the intermediate open conformation, while subunit B in state 2 best describes the intermediate closed conformation.
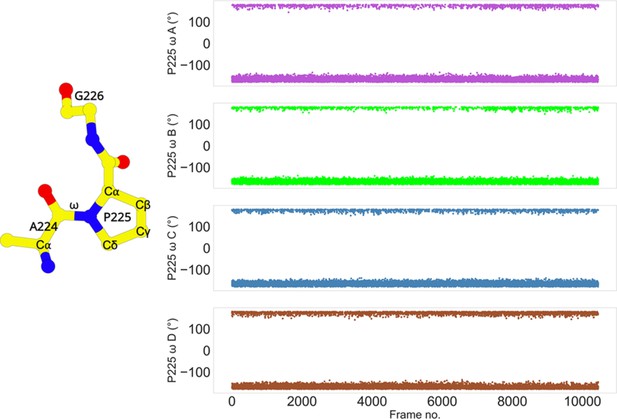
Trans conformation of P225.
The cis/trans conformation of a peptide bond is determined by the torsion of the ω angle. The ω torsion angle is close to 0° in the cis configuration, or ca. 180° in the trans configuration. The ω angle of P225 in each subunit transitions between 180° and −180°. The stride is 100 frames.

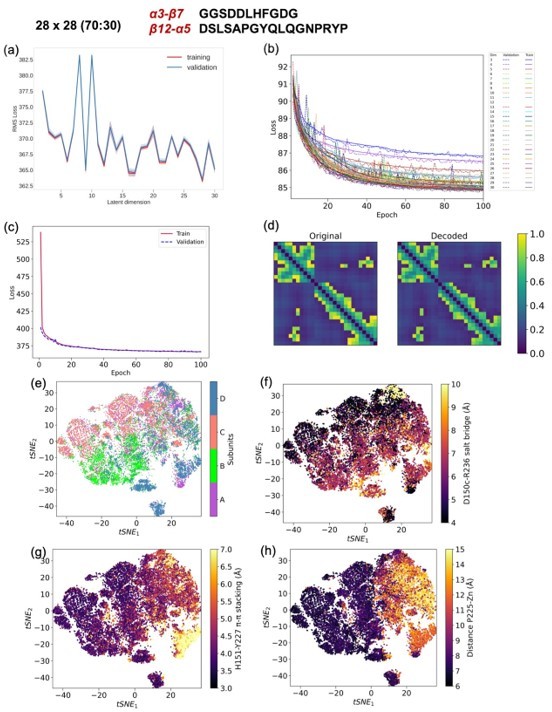
CVAE based deep learning analysis from a 28x28 matrix with 80 (training):20 (validation) data split.
(a) The training and validation loss plot as assessed over consecutive epochs at various reduced dimensions. (b) Validation loss during CVAE implementation is plotted at different latent dimensions for determining optimum values of the low dimension. The loss was calculated over 100 epochs from dimensions 3-30. (c) The convergence of training and validation loss from dimension 29, as assessed over 100 epochs. (d) Comparison between original input data and reconstructed data (decoded) are illustrated to ensure no loss of essential information during the compression and reconstruction process. (e) Subunit representation in the tsne space. The (f)salt bridge interaction between D150c-R236, (g) H151-Y227 p-p stacking interaction and (h) distance between P225 and the centroid of Zn atoms.
CVAE based deep learning analysis from a 28x28 matrix with 70 (training):30 (validation) data split.
(a) The training and validation loss plot as assessed over consecutive epochs at various reduced dimensions. (b) Validation loss during CVAE implementation is plotted at different latent dimensions for determining optimum values of the low dimension. The loss was calculated over 100 epochs from dimensions 3-30. (c) The convergence of training and validation loss from dimension 28, as assessed over 100 epochs. (d) Comparison between original input data and reconstructed data (decoded) are illustrated to ensure no loss of essential information during the compression and reconstruction process. (e) Subunit representation in the tsne space. The (f) salt bridge interaction between D150c-R236, (g) H151-Y227 p-p stacking interaction and (h) distance between P225 and the centroid of Zn atoms.
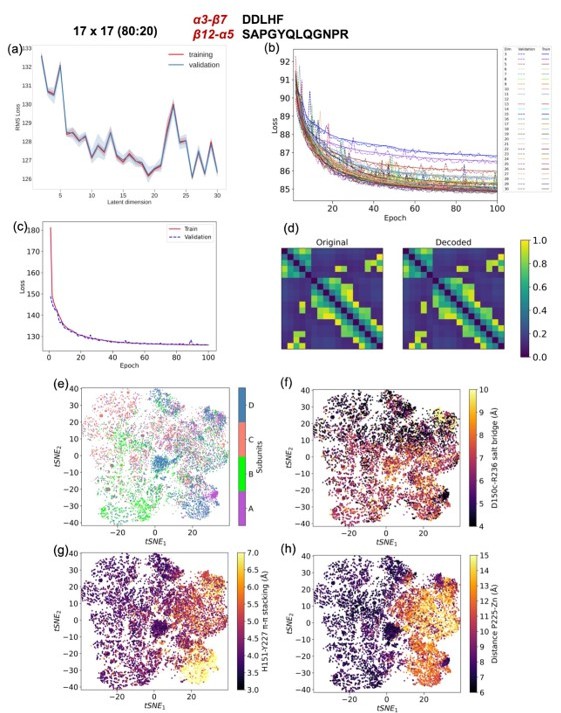
CVAE based deep learning analysis from a 17x17 matrix with 80 (training):20 (validation) data split.
(a) The training and validation loss plot as assessed over consecutive epochs at various reduced dimensions. (b) Validation loss during CVAE implementation is plotted at different latent dimensions for determining optimum values of the low dimension. The loss was calculated over 100 epochs from dimensions 3-30. (c) The convergence of training and validation loss from dimension 19, as assessed over 100 epochs. (d) Comparison between original input data and reconstructed data (decoded) are illustrated to ensure no loss of essential information during the compression and reconstruction process. (e) Subunit representation in the tsne space. The (f) salt bridge interaction between D150c-R236, (g) H151-Y227 p-p stacking interaction and (h) distance between P225 and the centroid of Zn atoms.
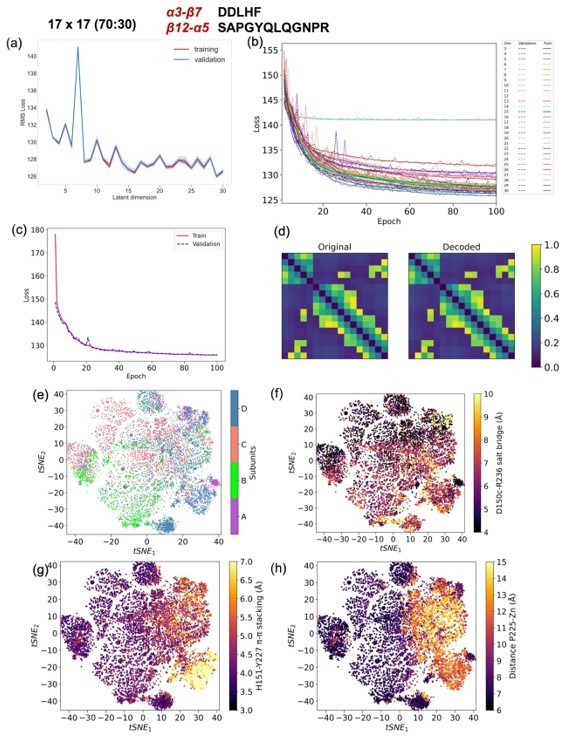
CVAE based deep learning analysis from a 17x17 matrix with 70 (training): 30 (validation) data split.
(a) The training and validation loss plot as assessed over consecutive epochs at various reduced dimensions. (b) Validation loss during CVAE implementation is plotted at different latent dimensions for determining optimum values of the low dimension. The loss was calculated over 100 epochs from dimensions 3-30. (c) The convergence of training and validation loss from dimension 29, as assessed over 100 epochs. (d) Comparison between original input data and reconstructed data (decoded) are illustrated to ensure no loss of essential information during the compression and reconstruction process. (e) Subunit representation in the tsne space. The (f) salt bridge interaction between D150c-R236, (g) H151-Y227 p-p stacking interaction and (h) distance between P225 and the centroid of Zn atoms.
Tables
Elongated loops.
| α3-β7 | GGSDDLHFGDGITYPPANAD Residues 149–171 |
| β12-α5 | ADSLSAPGYQLQGNPRYPH Residues 219–239 |
Susceptibility results of various β-lactams against L1 variants expressed in the pBC SK (+) vector in E. coli DH10B cells.
The reported values are the mode of at least three biological replicates.
| Strain | MIC value (mg/l)* | |||||
|---|---|---|---|---|---|---|
| CAZ | FEP | IMI | MER | TEB | TEB + CS319† | |
| DH10B pBC SK blaL1a | 256 | 2 | 16 | 16 | 32 | 2 |
| DH10B pBC SK blaL1a_H151V | 2 | 0.03 | 0.125 | 0.03 | ≤0.03 | ≤0.03 |
| DH10B pBC SK blaL1a_D150cV | 256 | 1 | 8 | 8 | 4 | 0.25 |
| DH10B pBC SK blaL1a_P225A | 64 | 0.125 | 0.25 | 2 | 0.125 | ≤0.03 |
| DH10B pBC SK blaL1a_R236A | 256 | 2 | 8 | 8 | 4 | 0.25 |
| DH10B pBC SK blaL1a_Y227A | 64 | 0.25 | 0.25 | 2 | 0.5 | ≤0.03 |
-
*
CAZ, ceftazidime; FEP, cefepime; IMI, imipenem; MER, meropenem; TEB, tebipenem.
-
†
CS319 was added to a final concentration of 100 mg/l.