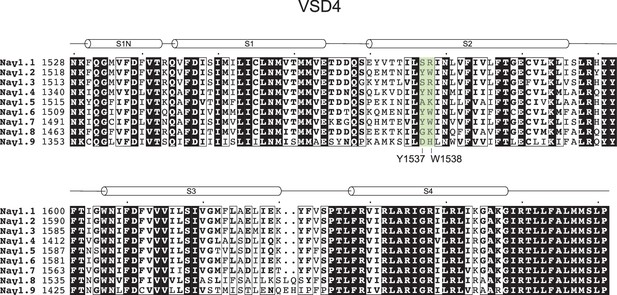
Cryo-EM reveals an unprecedented binding site for NaV1.7 inhibitors enabling rational design of potent hybrid inhibitors
Figures
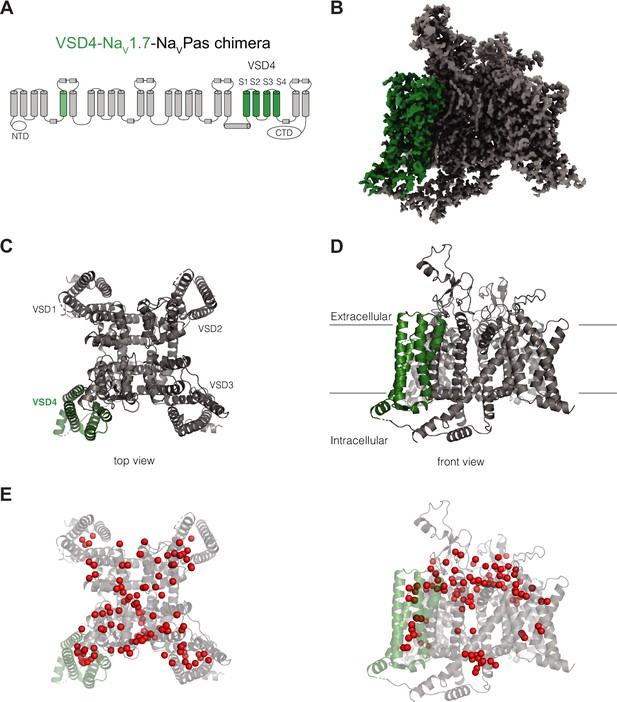
Structure of VSD4-NaV1.7-NaVPas.
(A) Schematic of the VSD4-NaV1.7-NaVPas channel. The portions humanized to the NaV1.7 sequence are shown in green. N-terminal domain (NTD) and CTD are indicated. (B) Side view of the single-particle cryogenic electron microscopy (cryo-EM) reconstruction of VSD4-NaV1.7-NaVPas channel. (C, D) Cartoon representations of the top and side views of VSD4-NaV1.7-NaVPas channel. Individual VSD domains are indicated. VSD4 is highlighted in green. (E) Localization of water molecules (in red) in the VSD4-NaV1.7-NaVPas channel structure. VSD4 is highlighted in green.
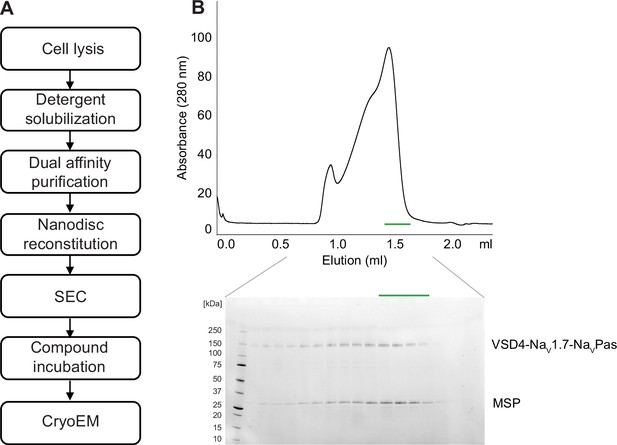
Purification of VSD4-NaV1.7-NaVPas channel.
(A) VSD4-NaV1.7-NaVPas channel protein expression and purification scheme. (B) Example size-exclusion chromatogram and SDS-PAGE of nanodisc-reconstituted VSD4-NaV1.7-NaVPas channel sample.
-
Figure 1—figure supplement 1—source data 1
Source data of VSD4-NaV1.7-NaVPas purification.
- https://cdn.elifesciences.org/articles/84151/elife-84151-fig1-figsupp1-data1-v2.zip
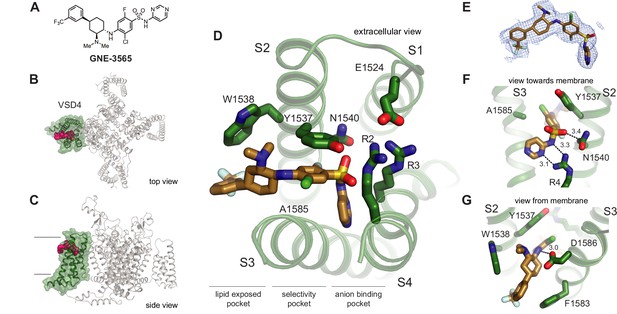
Structure of GNE-3565 bound to VSD4-NaV1.7-NaVPas.
(A) Chemical structure of arylsulfonamide GNE-3565. (B, C) Top and side views of VSD4-NaV1.7-NaVPas channel bound to GNE-3565. VSD4 is highlighted in green, GNE-3565 in magenta. (D) Extracellular view of VSD4-NaV1.7-NaVPas arylsulfonamide receptor site is shown with select side chains rendered as sticks. (E) The cryogenic electron microscopy (cryo-EM) map surrounding the ligand GNE-3565 is shown in mesh representation. (F) View toward the membrane highlighting key interactions with the GNE-3565 anionic group. (G) View from the membrane highlighting key interactions with the GNE-3565 central phenyl ring.
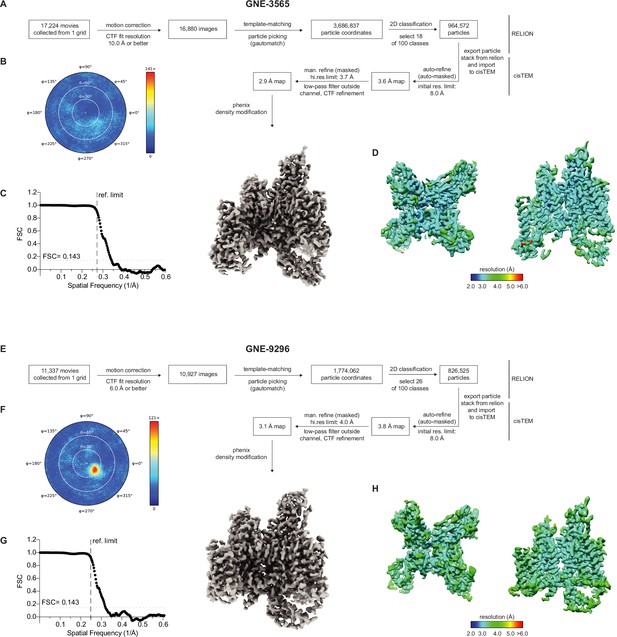
Cryo-EM processing workflow of GNE-3565 and GNE-9296.
(A) Data collection and processing workflow for VSD4-NaV1.7-NaVPas channel bound to GNE-3565. (B) Heat map representation of the distribution of assigned particle orientations. (C) Fourier shell corellation (FSC) between two half datasets yields a global resolution estimate of approximately 2.9 Å resolution from the refinement using an overall mask of the VSD4-NaV1.7-NaVPas channel bound to GNE-3565. (D) Local resolution of maps. (E) Data collection and processing workflow for VSD4-NaV1.7-NaVPas channel bound to GNE-9296 (compound 2). (F) Heat map representation of the distribution of assigned particle orientations. (G) FSC between two half datasets yields a global resolution estimate of approximately 3.1 Å resolution from the refinement using an overall mask of the VSD4-NaV1.7-NaVPas channel bound to GNE-9296 (compound 2). (H) Local resolution of maps.
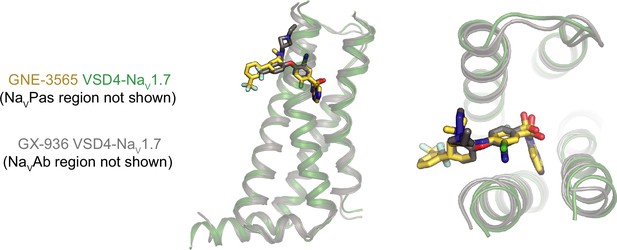
Comparison of VSD4-NaV1.7 bound to the arylsulfonamides GNE-3565 and GX-936 (PDB:5EK0).
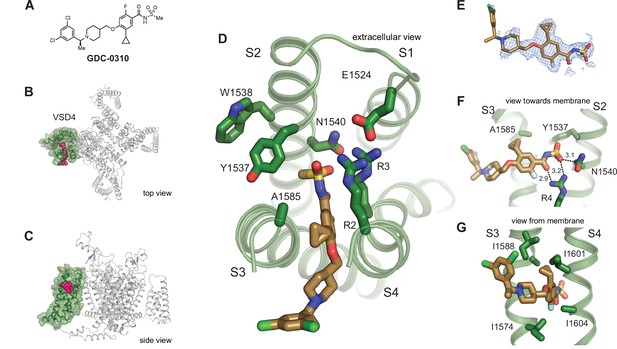
Structure of GDC-0310 bound to VSD4-NaV1.7-NaVPas.
(A) Chemical structure of acylsulfonamide GDC-0310. (B) Top and side views of VSD4-NaV1.7-NaVPas channel bound to GDC-0310. VSD4 is highlighted in green, GDC-0310 in magenta. (D) Extracellular view of VSD4-NaV1.7-NaVPas acylsulfonamide receptor site is shown with select side chains rendered as sticks. (E) The cryogenic electron microscopy (cryo-EM) map surrounding the ligand GDC-0310 is shown in mesh representation. (F) View toward the membrane highlighting key interactions with the GDC-0310 warhead. (G) View from the membrane highlighting van der Waals interactions with the GDC-0310 cyclopropyl substituent. The alpha-methylbenzylamine tail region sits almost entirely within the lipid bilayer.
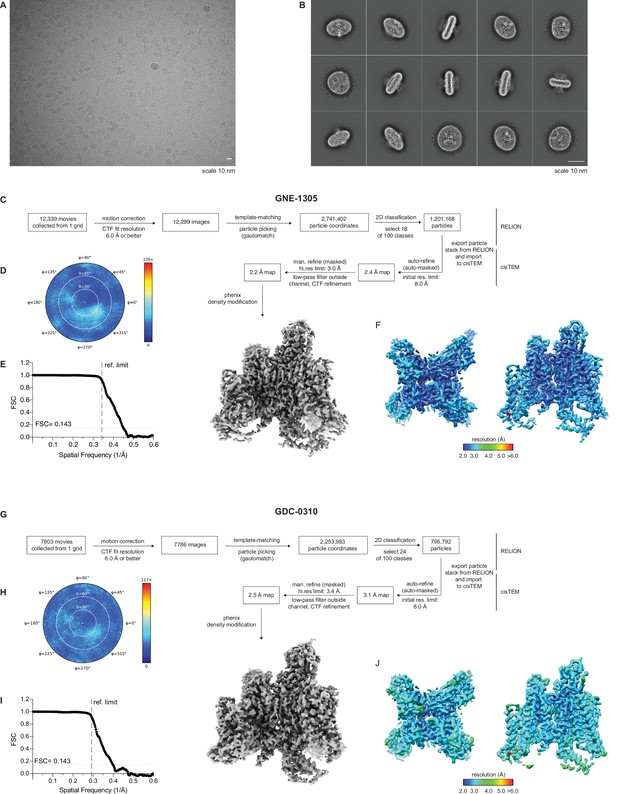
CyroEM processing workflow for GNE-1305 and GDC-0310.
(A) Example cryogenic electron microscopy (cryo-EM) micrograph image of the VSD4-NaV1.7-NaVPas channel. (B) Representative 2D-class averages of selected particles. (C) Data collection and processing workflow for VSD4-NaV1.7-NaVPas channel bound to compound 4. (D) Heat map representation of the distribution of assigned particle orientations. (E) FSC between two half datasets yields a global resolution estimate of approximately 2.2 Å resolution from the refinement using an overall mask of the VSD4-NaV1.7-NaVPas channel bound to GNE-1305 (compound 4) (F) Local resolution of maps. (G) Data collection and processing workflow for VSD4-NaV1.7-NaVPas channel bound to GDC-0310. (H) Heat map representation of the distribution of assigned particle orientations. (I) FSC between two half datasets yields a global resolution estimate of approximately 2.5 Å resolution from the refinement using an overall mask of the VSD4-NaV1.7-NaVPas channel bound to GDC-0310. (J) Local resolution of maps.
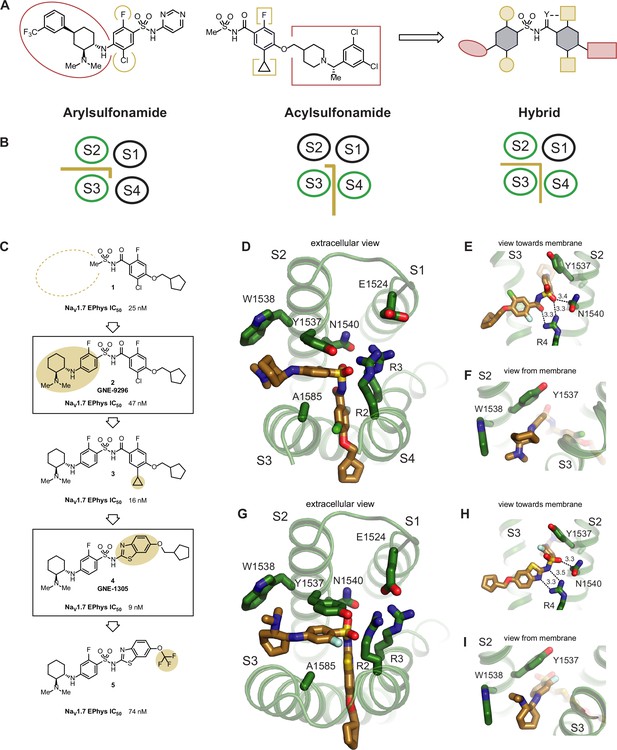
Structure-based design of potent hybrid inhibitors of NaV1.7.
(A) Illustration of hybrid molecule design approach. (B) Arylsulfonamide, acylsulfonamide, and hybrid molecule poses. (C) Hybridization strategy and molecule optimization. Highlighted are molecule 2 (GNE-9296) and molecule 4 (GNE-1305). (D) Extracellular view of VSD4-NaV1.7-NaVPas bound to the hybrid molecule 2 (GNE-9296). (E) View toward the membrane highlighting key interactions with the anionic group. (F) View from the membrane highlighting the lack of a stacking interaction between Y1537 and the phenyl ring. (G) Extracellular view of VSD4-NaV1.7-NaVPas bound to the hybrid molecule 4 (GNE-1305). (H) View toward the membrane highlighting key interactions with the anionic group. (I) View from the membrane highlighting the p-stacking interaction between Y1537 and the phenyl ring.
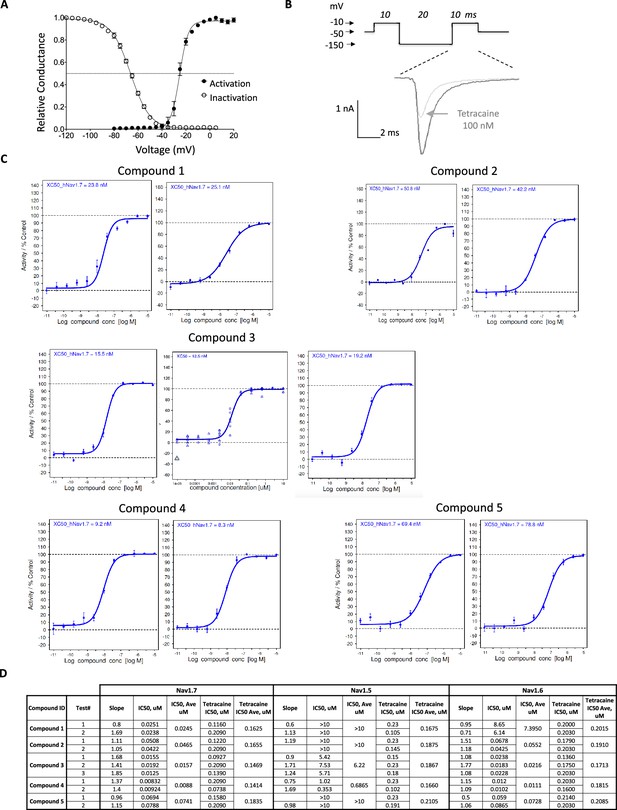
Biophysical and pharmacological characterization of human Nav1.7 channel by using Syncropatch384.
(A) Activation and inactivation curves of Nav1.7. Activation V1/2 = –25.2 mV (95% CI: –25.8 to –24.6 mV, n = 36). Inactivation V1/2 = –66.2 mV (95% CI: –66.6 to –65.7 mV, n = 153). (B). Pharmacology protocol and representative current traces for tetracaine block. Cells were held at –50 mV, pulsed to –10 mV for 10 ms, followed by a 20 ms pulse at –150 mV and 10 ms pulse to –10 mV. Currents elicited at the second –10 mV pulse were used to derive inhibition at inactivated states. Current traces showed 100 nM tetracaine block of Nav1.7 during a 7.5 min time period (from black to gray trace). (C) Representative dose responses of key compounds on human Nav1.7 channel. For each compound, 2–3 repeats were run, along with tetracaine control. (D) Summary of inhibition potency on Nav1.7, Nav1.5, and Nav1.6. Please note tetracaine was included as control for all experiments.
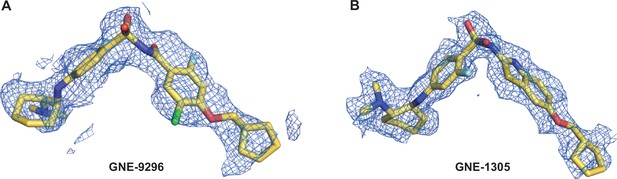
Density maps of GNE-9296 and GNE-1305.
(A) The cryogenic electron microscopy (cryo-EM) map surrounding the ligand GNE-9296 (compound 2) is shown in mesh representation. (B) The cryo-EM map surrounding the ligand GNE-1305 (compound 4) is shown in mesh representation.
Videos
Movie illustrating the quality of VSD4-NaV1.7-NaVPas channel bound to GNE-1305.
Several water molecules can be observed.
Additional files
-
Supplementary file 1
Experimental details for Cryo-EM, compound synthesis and characterization, and NaV subtype selectivity for selected NaV1.7 inhibitors.
(A) Cryogenic electron microscopy (cryo-EM) data collection, refinement and validation statistics. (B) NaV subtype selectivity for selected hybrid compounds.
- https://cdn.elifesciences.org/articles/84151/elife-84151-supp1-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84151/elife-84151-mdarchecklist1-v2.docx