A survey of open questions in adaptive therapy: Bridging mathematics and clinical translation
Figures
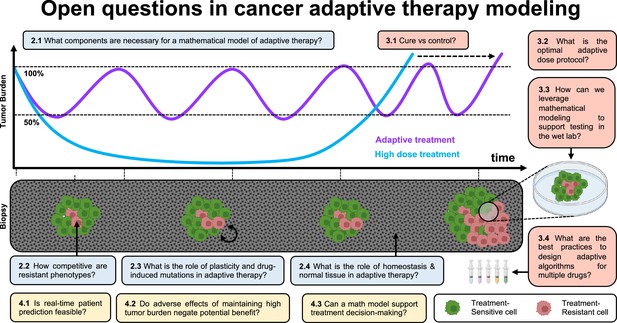
Open questions in adaptive cancer therapy modeling: schematic of tumor burden under maximum tolerable dose (blue) and adaptive dosing (purple), with corresponding biopsies.
Adaptive therapy is designed to exploit competition between treatment-sensitive (green) and resistant (red) cells to prolong the emergence of resistance. 11 questions representing future challenges in the field of adaptive therapy are shown, and answered within the text. Questions are color-coded by section: integrating the appropriate components into mathematical models (blue), design and validation of dosing protocols (red), and challenges and opportunities in clinical translation (yellow).
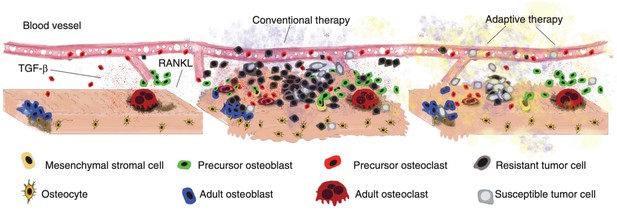
Disruption and restoration of tissue homeostasis.
Left: bone tissue homeostasis, including bone resorption by osteoclasts and osteoblasts. Middle: tumor cells cause disruption of homeostasis, leading to altered microenvironment factors. Conventional therapy leads to increasing tumor resistance. Right: evolution-based treatment strategies aim to restore some degree of homeostasis while allowing the tumor to remain sensitive to future treatment.
© 2017, Cold Spring Harbor Laboratory Press. Figure 2 is reproduced from Figure 2 of Basanta and Anderson, 2017 with permission from Cold Spring Harbor Laboratory Press. Copyright 2017 Cold Spring Harbor Laboratory Press; all rights reserved. It is not covered by the CC-BY 4.0 license and further reproduction of this panel would need permission from the copyright holder.
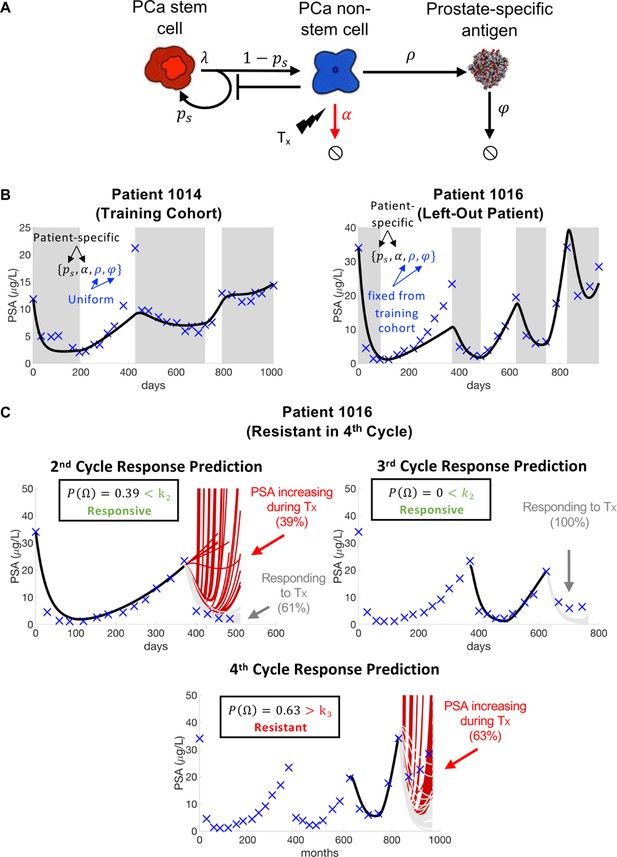
Model schematic, calibration, validation, and prediction.
Adapted from Figure 4 of Brady-Nicholls et al., 2021. (A) Model schematic of treatment-resistant stem cells, sensitive non-stem cells, and prostate-specific antigen interactions. (B) Model calibration (patient 1014) and validation (patient 1016). Nested optimization was used to determine the cohort uniform parameters and and the patient-specific parameters and for the training cohort. The uniform values were fixed in the testing cohort, and optimization was used to find the patient-specific parameters and . (C) Model predictions for patient 1016. The model predicted resistance in 39% of cycle 2 simulations and response in 100% of cycle 3 simulations. Cycle 4 predictions showed resistance in 63% of model simulations. Using cycle-specific cutoffs , and , the model correctly predicted that patient 1016 would continue to respond in cycles 2 and 3 but become resistant in cycle 4.