Phosphate starvation signaling increases mitochondrial membrane potential through respiration-independent mechanisms
Figures
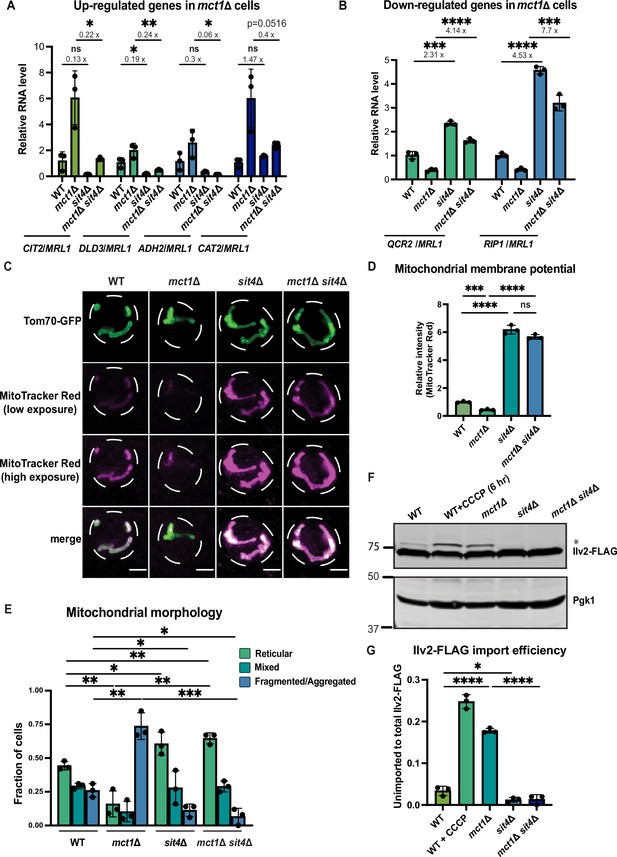
sit4Δ increases mitochondrial membrane potential in both wild-type and mct1Δ cells.
(A, B) Normalized gene expression of CIT2, DLD3, ADH2, CAT2, QCR2, and RIP1 measured 3 hr after switching from media containing 2% glucose as the sole carbon source to media containing 2% raffinose as the sole carbon source. Values were normalized to MRL1, a gene that was unchanged by deleting MCT1 in our RNA-seq dataset. n = 3. Fold changes are displayed. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant p>0.05; *p≤0.05; **p≤0.005; ***p≤0.0005; ****p≤0.0001 (C) Representative images of wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains expressing Tom70-GFP from its endogenous locus stained with MitoTracker Red. Scale bar represents 2 μm. (D) Normalized mitochondrial membrane potential of wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; ***p≤0.0005; ****p≤0.0001. (E) Quantification of the fraction of cells in (C) showing reticular, mixed, or fragmented/aggregated mitochondrial morphology based on Tom70-GFP signal. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; **p≤0.005; ***p≤0.0005. (F) Immunoblots of whole-cell lysates extracted from wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains expressing Ilv2 endogenously tagged with FLAG. As a control, wild-type (WT) cells were treated with 25 μM CCCP for 6 hr. * indicates unimported Ilv2-FLAG. Pgk1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 1—source data 1. (G) Normalized quantification of (F). Import efficiency is the ratio of unimported (*) to total abundance of Ilv2-FLAG. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; ****p≤0.0001. All original immunoblots used for quantification are displayed in Figure 1—source data 1.
-
Figure 1—source data 1
Source data and uncropped blots used to make Figure 1.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig1-data1-v2.zip

Genetic screen to identify SIT4 regulates nuclear responses induced in mct1Δ cells.
(A, B) Heat map visualizing selected gene expression between wild-type (WT) and mct1Δ using transcriptomics data from Berg et al., 2023. Gene expression was measured at 0, 3, and 12 hr after switching from glucose containing media to raffinose containing media. Genes in red and blue font were measured in the qPCR experiment shown in Figure 1A and B. All electron transport chain (ETC) and ATP synthase subunits and genes involved in acetyl-coA production are shown if they passed the detection and analysis criteria. (C) Schematic of the genetic screen. The coding sequences of CIT2 and BTT1 were replaced with Firefly luciferase and Renilla luciferase, respectively. CIT2 and BTT1 genes were restored at the HO locus to avoid any transcriptional alterations induced by their deletion. This query strain was then crossed with a deletion library, sporulated, and selected for double mutants in haploid (mct1ΔxxxΔ, where xxxΔ signifies the variable genes being deleted in the screen). Candidate genes were identified based on their ability to restore transcription back to wild-type levels of the Firefly:Renilla signal ratio. (D) Venn diagram of all the candidate mutants from the genetic screen. The genes listed inside the Venn diagram represent individual knockout strains. The genes (DLD3, CAT2, ADH2, and YAT1) above the Venn diagram indicate transcript that were used as indicators as measured by RT-qPCR. Genes listed outside the Venn diagram failed to reduce any of the indicator transcripts’ expression when deleted. (E) Individual colonies of mct1Δ cells were mated with rho0 cells and streaked onto a synthetic media supplemented with 2% glucose plate (SD) and then replica-plated onto a synthetic media supplemented with 2% glycerol plate (SG) to test for the presence of functional mtDNA. (F, G) Wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains expressing Tom70-GFP from its endogenous locus were stained with MitoTracker Red and imaged. 30–90 cells were captured for each analysis. Mitochondrial membrane potential was determined by quantification of the MitoTracker Red signal that co-localized with Tom70-GFP. Mitochondrial area was calculated by the percentage of Tom70-GFP signal in total cell area. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; *p≤0.05; **p≤0.005; ***p≤0.0005; ****p≤0.0001. (H) Normalized mitochondrial membrane potential of wild-type (WT), sit4Δ, and sit4Δ treated with different doses of CCCP quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. **p≤0.005; ****p≤0.0001. (I) Immunoblots of whole-cell lysates extracted from wild-type (WT) and sit4Δ strains expressing Ilv2 endogenously tagged with FLAG treated with indicated dosage of CCCP for 6 hr. * indicates unimported Ilv2-FLAG. Pgk1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 1—figure supplement 1—source data 1. (J) Normalized quantification of (I). Import efficiency is the ratio of unimported (*) to total abundance of Ilv2-FLAG. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant. All original immunoblots used for quantification are displayed in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Source data and uncropped blots used to make Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig1-figsupp1-data1-v2.zip
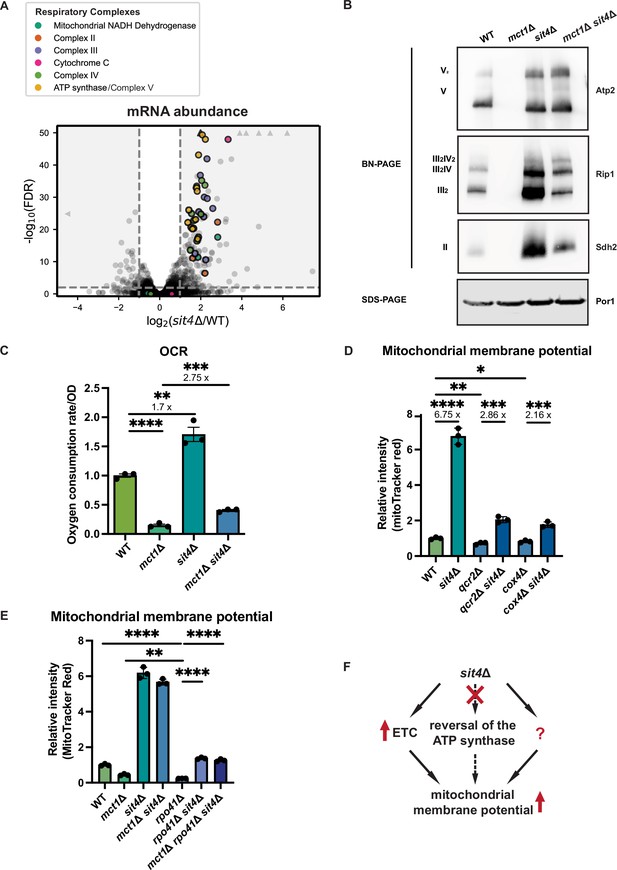
sit4Δ increases mitochondrial membrane potential through electron transport chain (ETC)-dependent and -independent mechanisms.
(A) Volcano plot of the transcriptomics data of sit4Δ vs. wild-type (WT) cells grown in synthetic media containing 2% glucose. All genes encoding components of the ETC and ATP synthase that were detected by RNA sequencing are highlighted and color-coded if they passed the detection and analysis criteria. Triangle indicates that the -log10 (FDR) exceeds 50. (B) Immunoblots of crude mitochondria extracted from wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains and separated on both BN-PAGE or SDS-PAGE. Membranes were blotted with indicated antibodies. Por1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 2—source data 1. (C) Normalized oxygen consumption rate (OCR) over optical density (OD) of the indicated strains grown in synthetic media containing 2% raffinose. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. **p≤0.005; ***p≤0.0005; ****p≤0.0001. (D, E) Normalized mitochondrial membrane potential of wild-type (WT), sit4Δ, qcr2Δ, qcr2Δ sit4Δ, cox4Δ, cox4Δ sit4Δ, mct1Δ, rpo41Δ, rpo41Δ sit4Δ, and mct1Δ rpo41Δ sit4Δ strains quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; **p≤0.005; ***p≤0.0005; ****p≤0.0001. (F) Schematic of mechanisms through which sit4Δ increases mitochondrial membrane potential. The mechanism (reversal of ATP synthase) that is theoretically possible but not utilized in sit4Δ cells is marked in dashed line.
-
Figure 2—source data 1
Source data and uncropped blots used to make Figure 2.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig2-data1-v2.zip
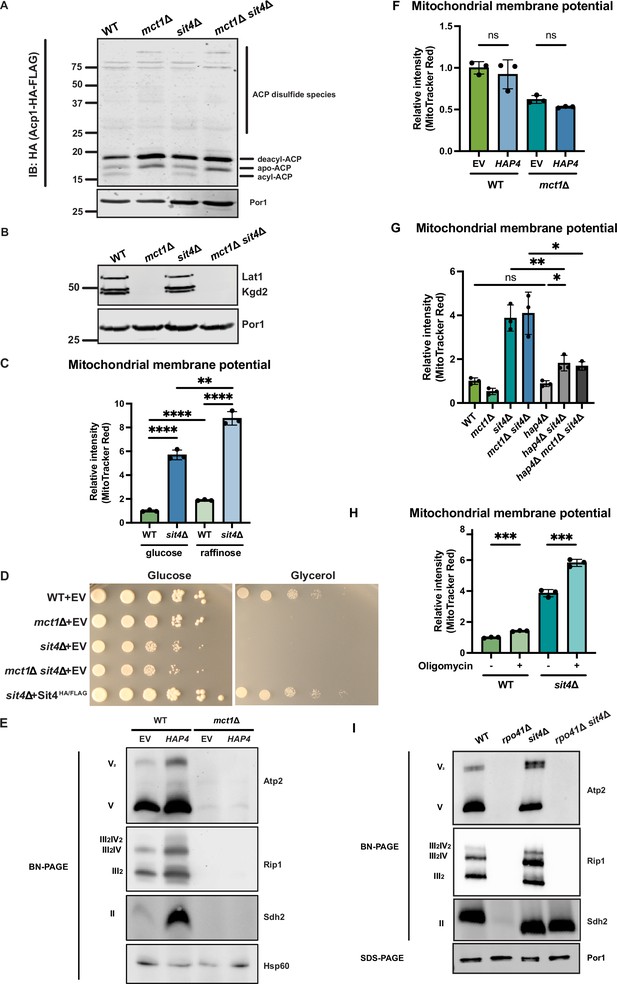
SIT4 deletion does not restore ACP acylation or respiratory growth in mct1Δ cells.
(A) Mitochondria were isolated from wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ cells expressing Acp1-HA-FLAG from a plasmid. Acp1-HA-FLAG was immunoprecipitated with HA agarose beads and the immunoprecipitate was separated by SDS-PAGE and immunoblotted with FLAG antibody. Por1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 2—figure supplement 1—source data 1. (B) Immunoblots of isolated mitochondria from wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ strains blotted with antibodies against lipoic acid or porin (Por1). Two bands around the molecular weight of Kgd2 were detected. Their exact identity is unclear but could indicate different post-translation modifications of the protein. Por1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 2—figure supplement 1—source data 1. (C) Normalized quantification of mitochondrial membrane potential measured by flow cytometry. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. **p≤0.005; ****p≤0.0001. (D) Spot tests measuring the growth rate of wild-type (WT), mct1Δ, sit4Δ, and mct1Δ sit4Δ on either glucose- or glycerol-containing synthetic media. (E) Immunoblots of crude mitochondria extracted from wild-type (WT) and mct1Δ strains overexpressing either empty vector (EV) or HAP4 and separated on BN-PAGE. Membranes were blotted with indicated antibodies. Hsp60 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 2—figure supplement 1—source data 1. (F) Normalized quantification of mitochondrial membrane potential measured by flow cytometry. n = 3. Error bars represent the SD. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons. ns = not significant; p>0.05. (G) Normalized quantification of mitochondrial membrane potential measured by flow cytometry. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; *p≤0.005; **p≤0.005. (H) Normalized quantification of mitochondrial membrane potential measured by flow cytometry. Wild-type (WT) and sit4Δ cells were treated with 5 μM oligomycin for 2 hr. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ***p≤0.0005. (I) Immunoblots of crude mitochondria extracted from wild-type (WT), rpo41Δ, sit4Δ, and rpo41Δ sit4Δ strains and separated on both BN-PAGE or SDS-PAGE. Membranes were blotted with indicated antibodies. Por1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Source data and uncropped blots used to make Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig2-figsupp1-data1-v2.zip
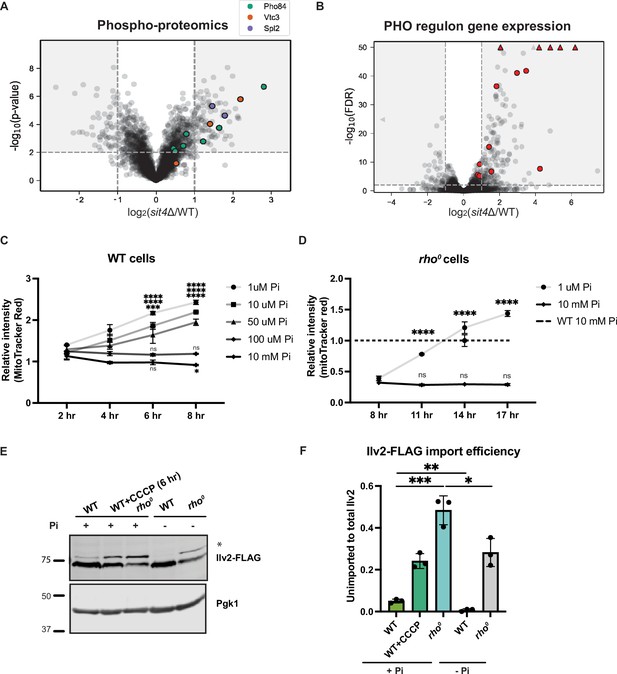
Phosphate starvation increases mitochondrial membrane potential through electron transport chain (ETC)-dependent and independent mechanisms.
(A) Volcano plot of phosphoproteomics data of sit4Δ vs. wild-type (WT) cells grown in synthetic media containing 2% glucose. Unique phosphorylation sites of Pho84 (green), Vtc3 (red), and Spl2 (purple) are highlighted. (B) Volcano plot of transcriptomics data of sit4Δ vs. wild-type (WT) cells grown in synthetic media containing 2% glucose. All PHO regulon targets that are detected by RNA sequencing are highlighted in red. Triangle indicates that the -log10 (FDR) exceeds 50. (C, D) Normalized time course of mitochondrial membrane potential in wild-type (WT) and rho0 strains measured by flow cytometry. The dashed line represents the membrane potential of wild-type (WT) cells grown in media containing 10 mM phosphate. n = 3. Fold changes are displayed. Error bars represent the SD. Statistical significance was determined using two-way ANOVA with Tukey’s multiple comparisons. ns = not significant; p>0.05; ***p≤0.0005; ****p≤0.0001. (E) Immunoblots of whole-cell lysates extracted from wild-type (WT) or rho0 cells expressing Ilv2 endogenously tagged with FLAG. Wild-type and rho0 cells were grown in media containing either 10 mM of phosphate (+Pi) or 1 μM of phosphate (-Pi) for 4 hr or overnight, respectively. As a control, wild-type (WT) cells were treated with 25 μM CCCP for 6 hr. * indicates unimported Ilv2-FLAG. Pgk1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 3—source data 1. (F) Normalized quantification of (E). Import efficiency is the ratio of unimported (*) to total abundance of Ilv2-FLAG. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; **p≤0.005; ***p≤0.0005. All original immunoblots used for quantification are displayed in Figure 3—source data 1.
-
Figure 3—source data 1
Source data and uncropped blots used to make Figure 3.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig3-data1-v2.zip
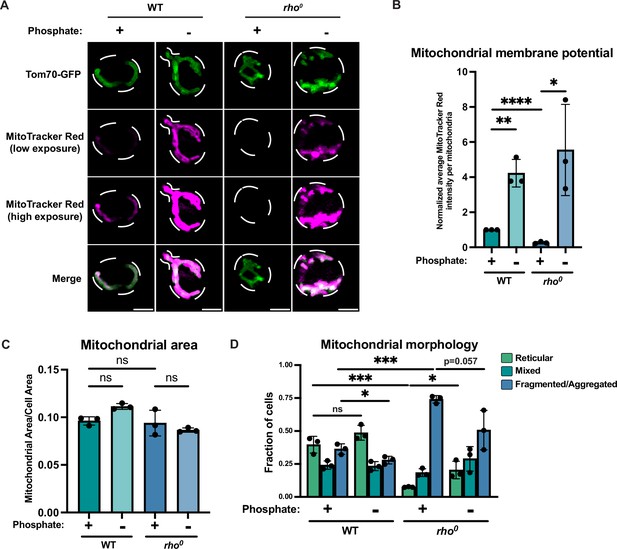
Phosphate depletion increases mitochondrial membrane potential in wild-type and rho0 cells.
(A) Wild-type (WT) cells expressing Tom70-GFP from its endogenous locus were grown in media containing high phosphate (10 mM Pi) or low phosphate (1 μM Pi) for 4 hr. rho0 cells expressing Tom70-GFP from its endogenous locus were grown in media containing high phosphate (+Pi, 10 mM Pi) or low phosphate (-Pi, 1 μM Pi) overnight. All cells were stained with MitoTracker Red and imaged. Representative images are shown. Scale bar represents 2 μm. (B) Quantification of (A). Mitochondrial membrane potential was determined by quantification of the MitoTracker Red signal that co-localized with Tom70-GFP. Mitochondrial area was calculated by the percentage of Tom70-GFP signal in total cell area. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; **p≤0.005; ****p≤0.0001. (C) Quantification of (A). Mitochondrial area was measured using Tom70-GFP signal. n = 3. Error bars represent the SD. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons. ns = not significant; p>0.05. (D) Quantification of the fraction of cells imaged in (A) showing reticular, mixed, or fragmented/aggregated mitochondrial morphology based on Tom70-GFP signal. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; *p≤0.05; ***p≤0.0005.
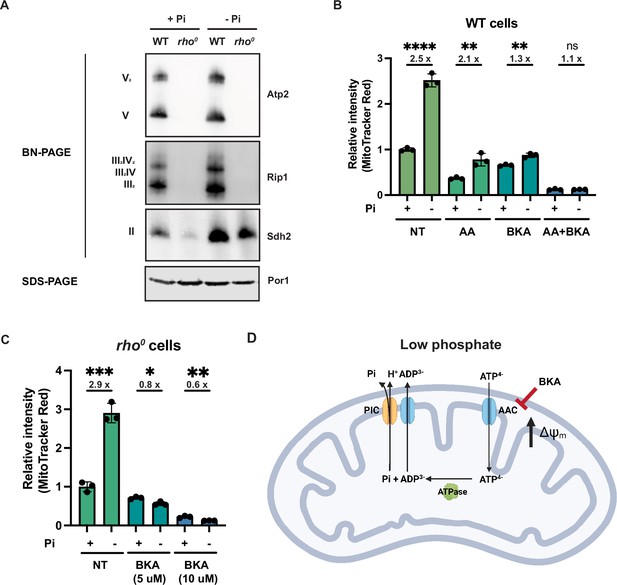
Phosphate depletion promotes mitochondrial membrane potential via ADP/ATP carrier in cells without electron transport chain (ETC) and ATP synthase.
(A) Wild-type (WT) and rho0 cells were grown in 10 mM (+Pi) or 1 μM (-Pi) phosphate-containing media for 4 hr or overnight, respectively. Crude mitochondria were extracted and separated by BN-PAGE or SDS-PAGE. Membranes were blotted with the indicated antibodies. Por1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 4—source data 1. (B) Wild-type (WT) cells were grown in 10 mM (+Pi) or 1 μM (-Pi) phosphate-containing media with or without drug treatment for 4 hr. Mitochondrial membrane potential was measured and quantified by flow cytometry. Fold changes are displayed. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; **p≤0.005; ****p≤0.0001. (C) rho0 cells were grown in 10 mM (+Pi) and 1 μM (-Pi) phosphate-containing media overnight and treated with or without bongkrekic acid (BKA) for 4 hr. Mitochondrial membrane potential was quantified by flow cytometry measurements of MitoTracker Red. Fold changes are displayed. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05; **p≤0.005; ***p≤0.0005. (D) Schematic of the mechanisms for increased mitochondrial membrane potential induced by phosphate depletion.
-
Figure 4—source data 1
Source data and uncropped blots used to make Figure 4.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig4-data1-v2.zip
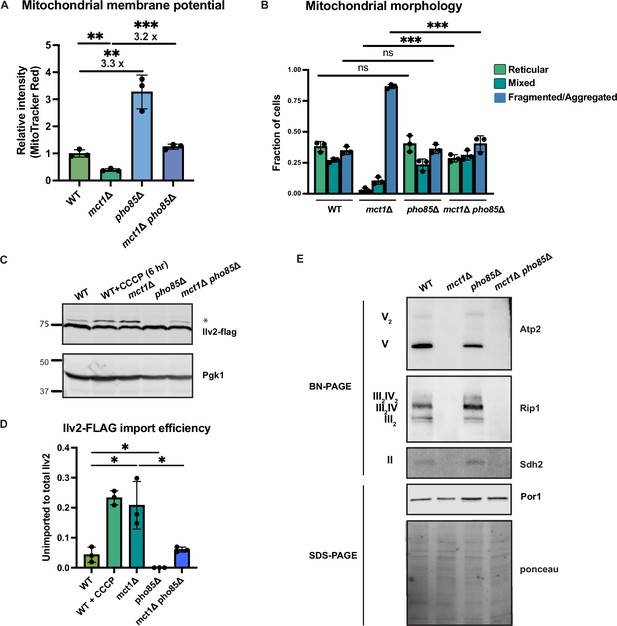
Activation of phosphate signaling increases mitochondrial membrane potential.
(A) Normalized mitochondrial membrane potential of wild-type (WT), mct1Δ, pho85Δ, and mct1Δ pho85Δ strains quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Fold changes are displayed. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. **p≤0.005; ***p≤0.0005. (B) Quantification of the fraction of cells in Figure 4B showing reticular, mixed, or fragmented/aggregated mitochondrial morphology based on Tom70-GFP signal. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; ***p≤0.0005. (C) Immunoblots of whole-cell lysates extracted from wild-type (WT), mct1Δ, pho85Δ, and mct1Δ pho85Δ cells expressing Ilv2 endogenously tagged with FLAG. As a control, wild-type (WT) cells were treated with 25 μM CCCP for 6 hr. * indicates unimported Ilv2-FLAG. Pgk1 was immunoblotted as a loading control. Original immunoblots are displayed in Figure 5—source data 1. (D) Normalized quantification of (C). Import efficiency is the ratio of unimported (*) to total abundance of Ilv2-FLAG. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05. All original immunoblots used for quantification are displayed in Figure 5—source data 1. (E) Immunoblots of crude mitochondria extracted from wild-type (WT), mct1Δ, pho85Δ, and mct1Δ pho85Δ cells and separated by BN-PAGE or SDS-PAGE. Membranes were blotted with indicated antibodies. The membrane was stained with Ponceau S and blotted with Por1 antibody as loading controls. Original immunoblots are displayed in Figure 5—source data 2.
-
Figure 5—source data 1
Source data and uncropped blots used to make Figure 5.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig5-data1-v2.zip
-
Figure 5—source data 2
Source data and uncropped blots used to make Figure 5.
- https://cdn.elifesciences.org/articles/84282/elife-84282-fig5-data2-v2.zip
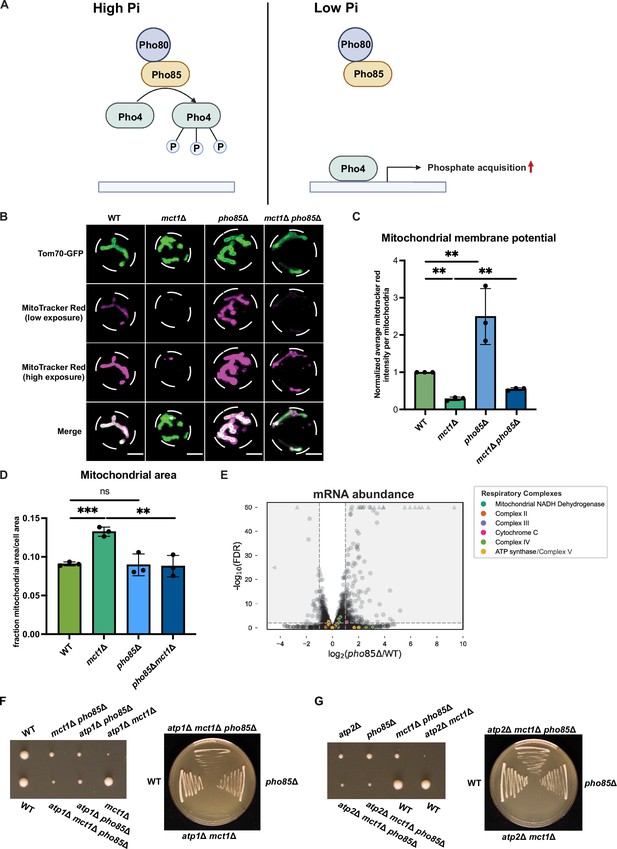
Phosphate starvation signaling increases mitochondrial membrane potential.
(A) Schematics of phosphate starvation signaling in yeast. (B) Representative images of wild-type (WT), mct1Δ, pho85Δ, and mct1Δ pho85Δ strains expressing Tom70-GFP from its endogenous locus stained with MitoTracker Red. Scale bar represents 2 μm. (C, D) Wild-type (WT), mct1Δ, pho85Δ, and mct1Δ pho85Δ strains expressing Tom70-GFP from its endogenous locus were stained with MitoTracker Red and imaged. 30–90 cells were captured for each analysis. Mitochondrial membrane potential was determined by quantification of the MitoTracker Red signal that co-localized with Tom70-GFP. Mitochondrial area was calculated by the percentage of Tom70-GFP signal in total cell area. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ns = not significant; p>0.05; **p≤0.005; ***p≤0.0005. (E) Volcano plot of transcriptomics data of pho85Δ vs. WT. All components of the ETC and ATP synthase genes that were detected by RNA-seq are highlighted and color-coded if they passed the detection and analysis criteria. Triangle indicates that the -log10 (FDR) exceeds 50. (F, G) Heterozygotic atp1Δ mct1Δ pho85Δ and atp2Δ mct1Δ pho85Δ cells were sporulated and dissected. Each haploid genotype was identified by their drug resistance. Individual colony was streaked on YPAD plate after tetrad dissection. Representative images are shown.
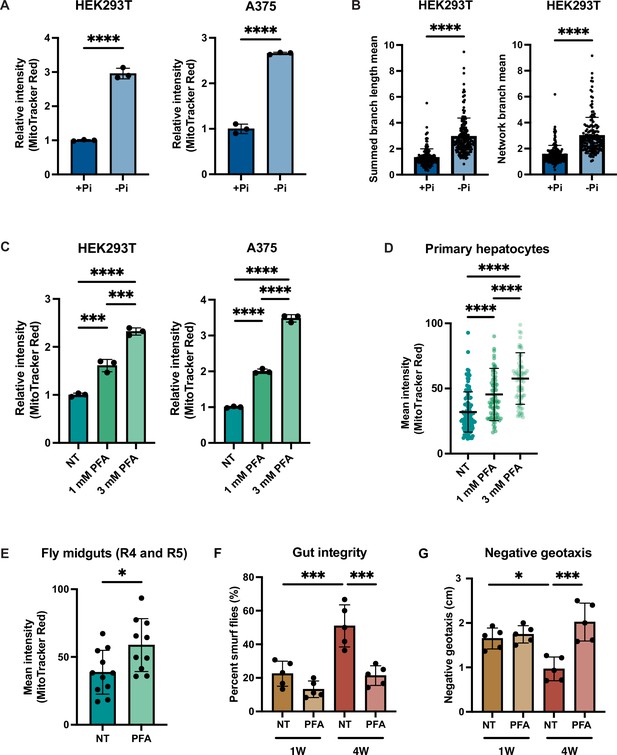
Phosphate depletion induces increased mitochondrial membrane potential in higher eukaryotic cells.
(A) The indicated cell lines were cultured with 1 mM (+Pi) or no phosphate (-Pi) for 3 d. Mitochondrial membrane potential was quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ****p≤0.0001. (B) Summed branch length mean and network branch mean were measured and calculated by Mitochondrial Network Analysis (MiNA). n = 3. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ****p≤0.0001. (C) HEK293T cells were treated with 0, 1, or 3 mM phosphonoformic acid (PFA) for 48 hr. Mitochondrial membrane potential was quantified by flow cytometry measurement of 10,000 cells stained with MitoTracker Red. n = 3. Error bars represent the SD. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons. ***p≤0.0005; ****p≤0.0001. (D) Primary hepatocytes were treated with 0, 1, or 3 mM PFA for 24 hr, and then stained with MitoTracker Red and imaged. The mean intensity of mitochondrial membrane potential was quantified by measurement of by the MitoTracker Red fluorescent signal from ~80 cells per condition. Error bars represent the SD. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons. ****p≤0.0001.(E) Three-week-old flies from the control group or from the experiment group treated with 1 mM PFA for 2 wk were dissected. Their midguts (R4 and R5 region) were stained with TMRE and the fluorescent signal was quantified by microscopic imaging. Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. *p≤0.05. (F, G) Three-day-old flies were maintained on food either containing no drug or 1 mM PFA for 1 or 4 wk. Smurf assays were repeated for five groups, each with six female and four male flies. RING assays were repeated for five groups, each with 15 female and 10 male flies. Error bars represent the SD. Statistical significance was determined using a one-way ANOVA with Tukey’s multiple comparisons. *p≤0.05; ***p≤0.0005.
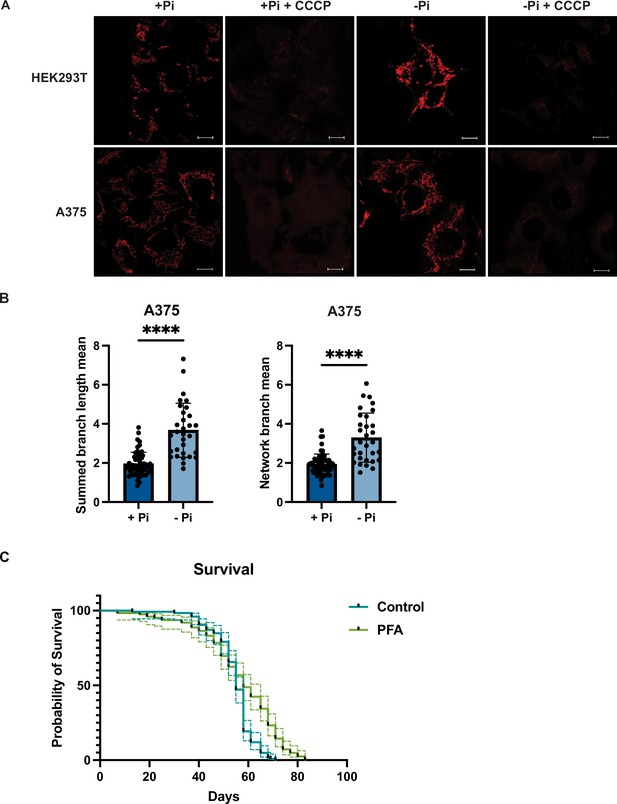
Phosphate depletion induces elevated mitochondrial membrane potential in mammalian cells.
(A) HEK293T and A375 cells were cultured in 1 mM (+Pi)- or no phosphate (-Pi)-containing media for 3 d. Cells were treated with 25 μM CCCP for 3 hr, stained with MitoTracker Red, and imaged. Representative images are shown. Scale bar represents 5 μM. (B) Summed branch lengths mean and network branch mean were measured and calculated by Mitochondrial Network Analysis (MiNA). Error bars represent the SD. Statistical significance was determined using an unpaired two-tailed t-test. ****p≤0.0001. (C) Three-day-old flies (15 females and 10 males in each group, five groups per condition) were maintained on food either containing no drug or 1 mM phosphonoformic acid (PFA). For 90 d, dead flies were counted every 3 d. 95% confidence interval was displayed in dashed lines. Statistical significance was determined using log-rank test. p<0.0001.
Tables
Summary of observations in sit4Δ cells or cells depleted with phosphate.
| Measurement | Background | Perturbation | |
|---|---|---|---|
| sit4Δ | Phosphate depletion | ||
| MMP | WT | High | High |
| mct1Δ | High | Higher than mct1Δ cells | |
| rpo41Δ or rho0 | Higher than rpo41Δ cells | Higher than rho0 cells | |
| Mitochondrial protein import efficiency | WT | More efficient | More efficient |
| mct1Δ | More efficient | More efficient than mct1Δ cells | |
| ETC | WT | Enriched | Unchanged |
| mct1Δ | More enriched than mct1Δ cells | Complex III, IV: remain absent Complex II: More enriched | |
Yeast strains used in this study.
| Genotype | Source | JRY identifier |
|---|---|---|
| WT (BY4741) | Van Vranken et al., 2018 | JRY 2884 |
| mct1::NatMX | Van Vranken et al., 2018 | JRY 2885 |
| can1::STE2p-Sp_HIS5 lyp1del btt1::Renilla-BTT1terminator-HygMX cit2::Firefly-CIT2terminator-Met15 ho::pr-cit2-term-pr-btt1-term-ura3 mct1::NatMX | This study | JRY 4614 |
| can1::STE2p-Sp_HIS5 lyp1del btt1::Renilla-BTT1terminator-HygMX cit2::Firefly-CIT2terminator-Met15 ho:: pr-cit2-term-pr-btt1-term-ura3 | This study | JRY 4616 |
| sit4::HygMX | This study | JRY 4144 |
| sit4::HygMX mct1::NatMX | This study | JRY 4145 |
| ILV2-FLAG::KanMX | Dr. Cory Dunn | JRY 4383/CDD1084 |
| mct1::NatMX ILV2-FLAG::KanMX | This study | JRY 4389 |
| sit4::HygMX ILV2-FLAG::KanMX | This study | JRY 4385 |
| sit4::HygMX mct1::NatMX ILV2-FLAG::KanMX | This study | JRY 4387 |
| qcr2::kanMX | This study | JRY 4556 |
| qcr2::kanMX sit4::HygMX | This study | JRY 4552 |
| cox4::kanMX | This study | JRY 4631 |
| cox4::kanMX sit4::HygMX | This study | JRY 4633 |
| rpo41::kanMX | This study | JRY 4719 |
| rpo41::kanMX sit4::HygMX | This study | JRY 4720 |
| rpo41::kanMX sit4::HygMX mct1::NatMX | This study | JRY 4721 |
| pho85::hisMX | This study | JRY 4715 |
| pho85::hisMX mct1::NatMX | This study | JRY 4716 |
| pho85::hisMX ILV2-FLAG::KanMX | This study | JRY 4743 |
| pho85::hisMX mct1::NatMX ILV2-FLAG::KanMX | This study | JRY 4744 |
| rho0 | This study | JRY 4941 |
| Tom70-yeGFP::hisMX | This study | JRY 7502 |
| Tom70-yeGFP::hisMX mct1::NatMX | This study | JRY 7504 |
| Tom70-yeGFP::hisMX sit4::HygMX | This study | JRY 7506 |
| Tom70-yeGFP::hisMX sit4::HygMX mct1::NatMX | This study | JRY 7508 |
| Tom70-yeGFP::hisMX rho0 | This study | JRY 7510 |
| Tom70-yeGFP::KanMX mct1::NatMX | This study | JRY 7514 |
| Tom70-yeGFP::KanMX pho85::hisMX | This study | JRY 7515 |
| Tom70-yeGFP::KanMX pho85::hisMX mct1::NatMX | This study | JRY 7516 |
Antibodies used in this study.
| Antibodies | Source |
|---|---|
| FLAG epitope | Sigma-Aldrich, F7425 |
| Sdh2 | Dr. Dennis Winge |
| Rip1 | Dr. Dennis Winge |
| Atp2 | Dr. Dennis Winge |
| Por1 | Abcam, ab110326 |
| Lipoic acid | Abcam, ab58724 |
| Pgk1 | Abcam, ab113687 |
Plasmids used in this study.
| Plasmids | Source |
|---|---|
| pRS416 Acp1-HA-FLAG | Van Vranken et al., 2018 |
| pRS416 Sit4-HA-FLAG | This study |
Chemicals and commercial kits used in this study.
| Chemicals | Source |
|---|---|
| sodium phosphonoformate tribasic hexahydrate | Sigma-Aldrich, P6801 |
| b-mercaptoethanol | Sigma-Aldrich, M6250 |
| Digitonin special-grade (water-soluble) | Gold Biotechnology, D-180 |
| Lyticase from Anthrobacter luteus | Sigma-Aldrich, L4025 |
| Protease inhibitor cocktail (yeast) | Sigma-Aldrich, P8215 |
| B-ethylmaleimide | Sigma-Aldrich, E3876 |
| anti-HA antibody-conjugated agarose | Sigma-Aldrich, A2095 |
| NativePAGE 20× running buffer | Invitrogen, BN2001 |
| NativePAGE 20× cathode buffer additive | Invitrogen, BN2002 |
| NativePAGE sample buffer (4×) | Invitrogen, BN20032 |
| NativePAGE 5% G-250 sample additive | Invitrogen, BN20041 |
| Ponceau S solution | Sigma-Aldrich, P7170 |
| Antimycin A | Sigma-Aldrich, A8674 |
| Bongkretic acid | Sigma-Aldrich, B6179 |
| MitoTracker Red CMXROS | Invitrogen Life, M7512 |
| CCCP | Sigma-Aldrich, C2759 |
| TMRE | Invitrogen, T669 |
| Hoechst 33342 | Thermo Scientific, 62249 |
| Bromophenol blue | Sigma-Aldrich, 114391 |
| Pierce BCA protein assay kit | Thermo Scientific, 23225 |
| Direct-zol RNA isolation kit | Zymo Research, R2050 |
| TURBO Dnase free kit | Invitrogen Life, AM1907 |
| LightCycler 480 SYBR Green I Master | Roche Life Science, 04707516001 |
| SuperSignal West Femto Max Sensitivity Substrate | Thermo Scientific, 34096 |
Additional files
-
Supplementary file 1
Primers used to create yeast strains.
- https://cdn.elifesciences.org/articles/84282/elife-84282-supp1-v2.xlsx
-
Supplementary file 2
RNA-sequencing results.
- https://cdn.elifesciences.org/articles/84282/elife-84282-supp2-v2.xlsx
-
Supplementary file 3
Phosphoproteomics results.
- https://cdn.elifesciences.org/articles/84282/elife-84282-supp3-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84282/elife-84282-mdarchecklist1-v2.pdf





