Chloride ions evoke taste sensations by binding to the extracellular ligand-binding domain of sweet/umami taste receptors
Figures
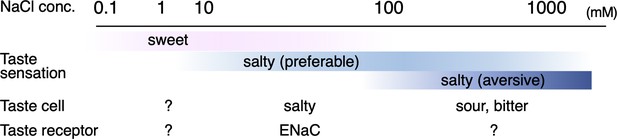
Salt taste sensation.
Approximate concentration ranges of salt taste perceptions in humans (Bartoshuk et al., 1978) and qualities of taste sensation with known cells and receptors responsible for their sensing are summarized.
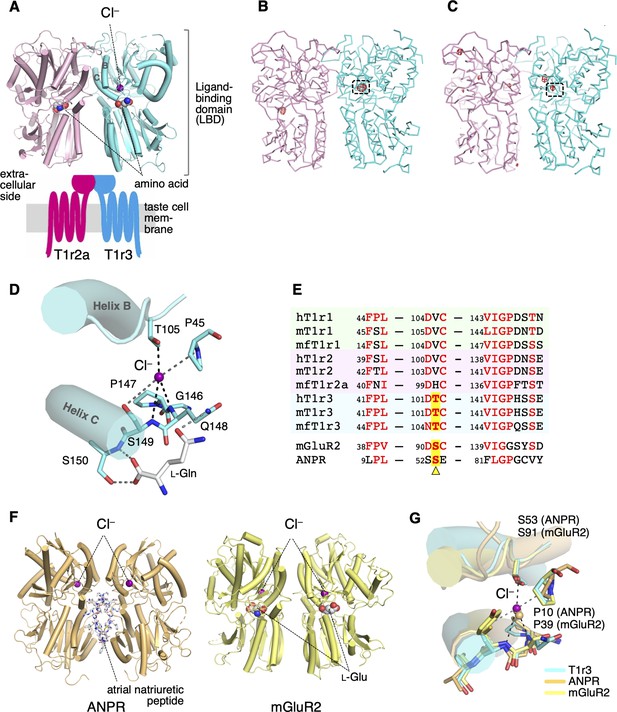
Cl−-binding sites in the medaka fish taste receptor T1r2a/T1r3LBD.
(A) Schematic drawing of the overall architecture of T1r2a/T1r3. The crystal structure (PDB ID: 5X2M) (Nuemket et al., 2017) is shown at the ligand-binding domain (LBD) region, and helices B and C in T1r3 are labeled. (B) Anomalous difference Fourier map (4.5 σ, red) of the Br−-substituted T1r2a/T1r3LBD crystal. (C) Anomalous difference Fourier map (4.5 σ, red) of the Cl−-bound T1r2a/T1r3LBD crystal derived from the diffraction data collected at the wavelength of 2.7 Å. In panels B and C, the site originally identified the Cl− binding was framed. (D) A close-up view of the Cl−-binding site in T1r3LBD in the Cl−-bound T1r2a/T1r3LBD (PDB ID: 5X2M). (E) Amino acid sequence alignment of T1r proteins and the related receptors at the Cl−-binding site. The ‘h’, ‘m’, and ‘mf’ prefixes to T1rs indicate human, mouse, and medaka fish, respectively. The position corresponding to Thr105 in T1r3 from medaka fish is highlighted. (F) The structures of atrial natriuretic peptide receptor (ANPR) (PDB ID: 1T34, left) (Ogawa et al., 2004) and mGluR2 (PDB ID: 5CNI, right) (Monn et al., 2015a) bound with Cl−. (G) Superposition of the Cl−-binding site in T1r3, ANPR, and mGluR2.
-
Figure 2—source data 1
The anomalous difference Fourier maps shown in Figure 2B, C.
The structure factor files and the coordinate files used for anomalous difference Fourier calculation, and the resultant maps were included.
- https://cdn.elifesciences.org/articles/84291/elife-84291-fig2-data1-v1.zip
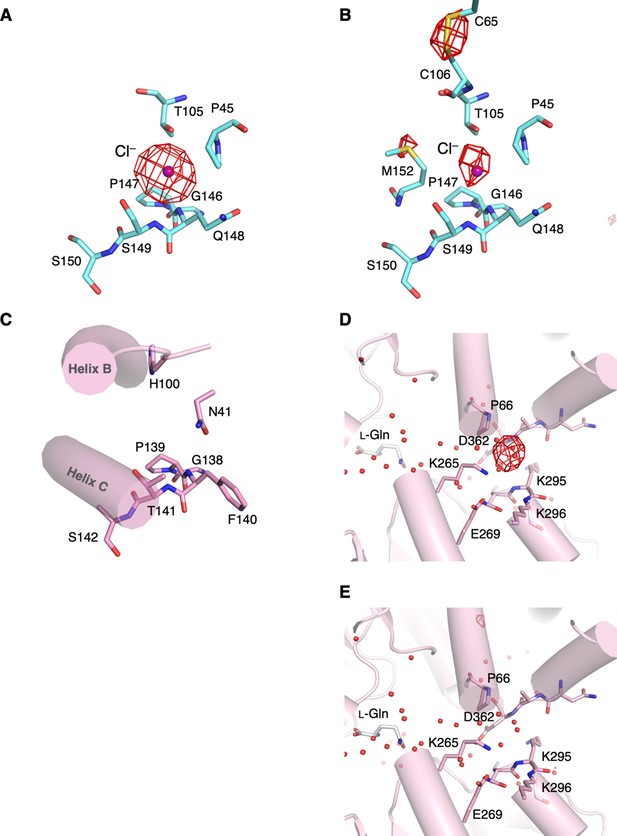
The structure of the regions relating to the Cl−-binding site in medaka fish T1r2a/T1r3LBD.
(A) The Br−-binding site in T1r3. The anomalous difference Fourier map (4.5 σ, red) of the Br−-substituted crystal is overlayed to the Cl−-bound T1r2a/T1r3LBD structure (PDB ID: 5X2M). (B) The Cl−-binding site in T1r3. The anomalous difference Fourier map (4.5 σ, red) calculated from the data collected at the wavelength of 2.7 Å is overlayed. (C) The region in T1r2a, which corresponds to the Cl−-binding site in T1r3, in the Cl−-bound T1r2a/T1r3LBD. (D) The Br−-binding site in T1r2a. The anomalous difference Fourier map (4.5 σ, red) of the Br--substituted crystal is overlayed to the Cl−-bound T1r2a/T1r3LBD structure. (E) The Br−-binding site in T1r2a. The 2.7 Å anomalous difference Fourier map (4.5 σ, red) of the Cl−-bound T1r2a/T1r3LBD, which is the same map shown in panel B, is overlayed to the Br−-binding site in T1r2a. No significant peak was observed.
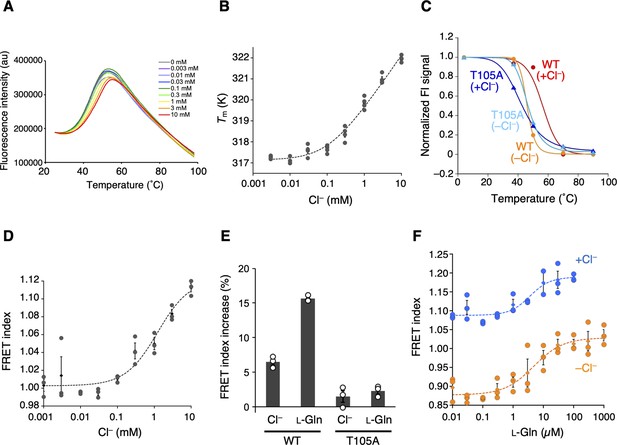
The Cl−-binding properties of T1r2a/T1r3LBD.
(A) Representative thermal melt curves of T1r2a/T1r3LBD in the presence of 0.003–10 mM concentrations of Cl− measured using differential scanning fluorimetry (DSF). (B) Dose-dependent Tm changes of T1r2a/T1r3LBD by addition of Cl− (n = 4). (C) Thermal melting curves of wild-type (WT) and the T1r3-T105A mutant of T1r2a/T1r3LBD in the presence and absence of Cl−, analyzed by fluorescence-detection size-exclusion chromatography-based thermostability (FSEC-TS) assay (n = 1). (D) Dose-dependent Förster resonance energy transfer (FRET) signal changes of the T1r2aLBD-Cerulean and T1r3LBD-Venus heterodimer by addition of Cl− (n = 3). (E) FRET index increases by adding 10 mM Cl− or 1 mM l-glutamine to the WT or T1r3-T105A mutant T1r2aLBD-Cerulean/T1r3LBD-Venus heterodimer relative to that in the absence of any ligand in the absence of Cl− (n = 3). (F) Dose-dependent FRET signal changes of the T1r2aLBD-Cerulean and T1r3LBD-Venus heterodimer induced by the addition of l-glutamine in the presence and absence of Cl− (n = 3). The experiments were performed two (panels A, B, D, E), three (C and WT), four (F and +Cl− condition), or one (C and mutant; F and −Cl− condition) time(s), and the results from one representative experiment are shown with numbers of technical replicates. Data points represent mean (panels B, D, F: diamonds; panel E: bars) ± standard error of the mean (SEM).
-
Figure 3—source data 1
Excel file with numerical data used for Figure 3.
- https://cdn.elifesciences.org/articles/84291/elife-84291-fig3-data1-v1.xlsx
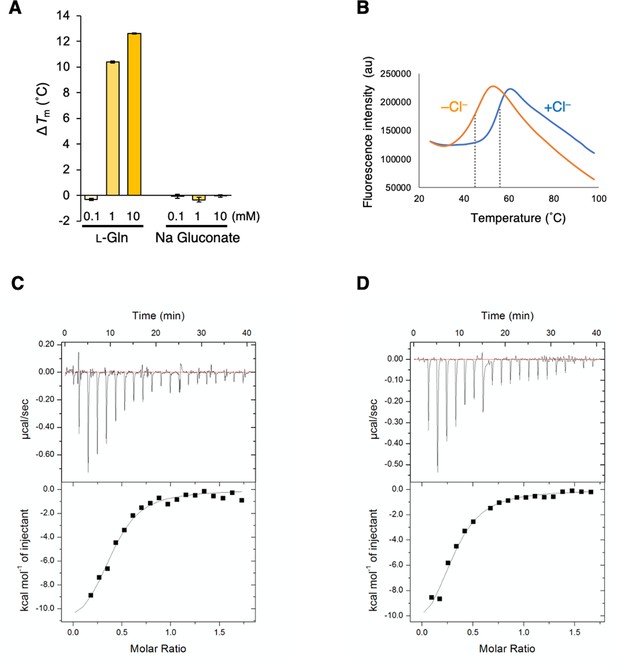
The properties of T1r2a/T1r3LBD in the presence and absence of Cl−.
(A) Binding analysis of gluconate by differential scanning fluorimetry (DSF). 0.1, 1, and 10 mM of l-glutamine, as a representative ligand, and sodium gluconate was added to T1r2a/T1r3LBD in 20 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesufonic acid (HEPES)–NaOH, 300 mM NaCl, pH 7.5. The mean increases of Tm (ΔTm) from that in the absence of a ligand (54.1°C, measured on the same sample, n = 2) are plotted. Error bars are in standard error of the mean (SEM) (n = 4). (B) Representative thermal melt curves of T1r2a/T1r3LBD in the presence or absence of Cl−, measured by DSF in the same condition in Figure 3C, and the panels C, D in this figure. (C, D) The l-glutamine binding to T1r2a/T1r3LBD was measured by isothermal titration calorimetry. The upper and lower panels show the raw data and integrated heat signals upon l-glutamine injection to T1r2a/T1r3LBD in the presence (C) and absence (D) of Cl−. The binding isotherms were fitted assuming 1 ligand: 1 heterodimer binding. The experiments were performed one (panel A), two (B), four (C), or five (D) time(s), and the results from one representative experiment are shown with numbers of technical replicates.
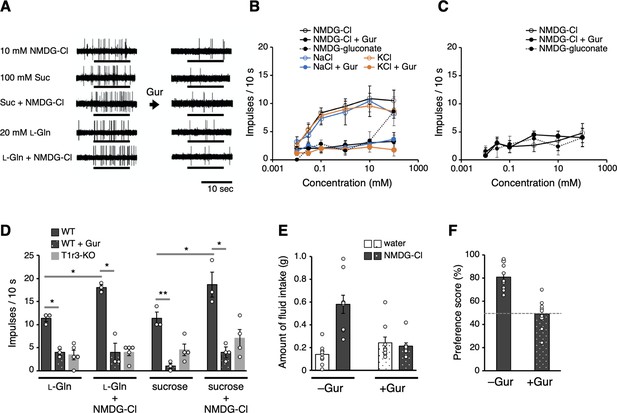
Electrophysiological and behavioral analyses of the T1r-mediated Cl− responses in mouse.
(A–D) Results of single fiber recordings from the mouse chorda tympani nerve. (A) Representative recordings of single fibers that connect to T1r-expressing taste cells. The stimuli were 10 mM NMDG-Cl, 100 mM sucrose, 100 mM sucrose + 10 mM NMDG-Cl, 20 mM l-glutamine, or 20 mM l-glutamine + 10 mM NMDG-Cl. Lines indicate the application of stimuli to the tongue. All the responses were suppressed by lingual treatment with a T1r blocker, Gur (right). (B) Impulse frequencies in response to the concentration series of NMDG-Cl, NaCl, or KCl before and after Gur treatment in wild-type (WT) mice. Responses to NMDG-gluconate are also shown. The mean number of net impulses per 10 s (mean response) ± standard error of the mean (SEM) in Gur-sensitive fibers (n = 5–6 from six mice). (C) Impulse frequencies in response to the concentration series of NMDG-Cl before and after Gur treatment were measured in T1r3-KO mice (n = 4–5 from three mice). Responses to NMDG-gluconate are also shown. (D) Impulse frequencies to 20 mM l-glutamine or 100 mM sucrose in the absence or presence of 10 mM NMDG-Cl before and after Gur treatment. Responses to 20 mM l-glutamine or 100 mM sucrose by T1r3-KO mouse are also shown. Values are mean (bars) ± SEM (n = 3–5 from three mice each). *, **: paired t-test; *p < 0.05 and **p < 0.01. (E) Amount of fluid intake for water and 10 mM NMDG-Cl in the two-bottle preference tests. Values are mean (bars) ± SEM (n = 9 mice). (F) NMDG-Cl intake shown in (E) normalized to water intake (preference score) in the two-bottle preference tests. A score >50% indicates that the taste solution was preferred over water.
-
Figure 4—source data 1
Excel file with numerical data used for Figure 4.
- https://cdn.elifesciences.org/articles/84291/elife-84291-fig4-data1-v1.xlsx
Tables
X-ray data collection statistics of T1r2a/T1r3LBD–Fab16A complex.
| Br− bound | Cl− bound | |
|---|---|---|
| Beamline | SPring-8 BL41XU | Photon Factory BL-1A |
| Detector | PILATUS6M | EIGER X4M |
| Wavelength (Å) | 0.9194 | 2.7 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a (Å) | 102.8 | 102.8 |
| b (Å) | 121.6 | 120.8 |
| c (Å) | 129.9 | 129.1 |
| Resolution (Å) | 50–3.41 (3.43–3.41) | 49.8–3.32 (3.33–3.32) |
| Rsym (%)* | 0.094 (0.808) | 0.089 (0.865) |
| I/σ(I)* | 16.4 (2.3) | 15.1 (2.5) |
| Completeness (%)* | 99.8 (99.2) | 99.6 (97.8) |
| Redundancy* | 7.0 (7.0) | 6.9 (6.8) |
-
*
Values in parentheses refer to data in the highest resolution shells.
Properties of the Cl− binding to T1r2a/T1r3LBD.
| Cl− binding, DSF (n = 4) | ||
|---|---|---|
| Kd-app (mM) | 0.111 ± 0.046 | |
| Protein thermal stability, DSF (n = 6) | ||
| Condition | +Cl− | −Cl− |
| Melting temperature (°C) | 55.2 ± 0.03 | 46.6 ± 0.06 |
| Protein thermal stability, FSEC-TS | ||
| Wild-type T1r2a/T1r3LBD | ||
| Condition | +Cl− | −Cl− |
| Melting temperature (°C) | 56.4 ± 5.1 | 46.0 ± 0.3 |
| Mutant T1r2a/T1r3-T105ALBD | ||
| Condition | +Cl− | −Cl− |
| Melting temperature (°C) | 42.7 ± 0.1 | 46.7 ± 0.7 |
| Cl− binding, FRET (n = 3) | ||
| FRET index minimum | 1.00 ± 0.005 | |
| FRET index change | 0.119 ± 0.014 | |
| EC50 (mM)* | 1.23 ± 0.53 | |
| l-Glutamine binding, FRET (n = 3) | ||
| Condition | +Cl− | −Cl− |
| FRET index minimum | 1.09 ± 0.01 | 0.88 ± 0.01 |
| FRET index change | 0.10 ± 0.02 | 0.15 ± 0.02 |
| EC50 (µM) | 3.59 ± 1.74 | 4.78 ± 1.41 |
| Hill coefficient | 1.31 ± 0.74 | 0.90 ± 0.22 |
| l-Glutamine binding, ITC | ||
| Condition | +Cl− | −Cl− |
| N (sites) | 0.389 ± 0.028 | 0.303 ± 0.023 |
| Ka (M−1) [converted to Kd (µM)] | (2.85 ± 0.65) × 105 [3.51] | (2.12 ± 0.46) × 105 [4.72] |
| ΔH (kcal/mol) | −12.3 ± 1.2 | −12.9 ± 1.3 |
| ΔS (cal/mol/deg) | −16.3 | −18.8 |
-
*
Hill coefficient was fixed to 1 for fitting.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, male) | C57BL/6JCrj | Charles River Japan | ||
| Genetic reagent (Mus musculus) | T1r3GFP-KO | This study | See Materials and methods, ‘Single fiber recording from mouse chorda tympani (CT) nerve’ subsection | |
| Cell line (Drosophila melanogaster) | S2 | Invitrogen | Cat # R69007 | |
| Cell line (Drosophila melanogaster) | S2, high-expression clone for medaka T1r2a/T1r3LBD | DOI:10.1038/ncomms15530 | ||
| Cell line (Drosophila melanogaster) | S2, high-expression clone for medaka T1r2aLBD-Cerulean/T1r3LBD-Venus | DOI:10.1038/ncomms15530 | ||
| Antibody | Anti-medaka T1r2a, clone 16 A (mouse monoclonal) | DOI:10.1038/ncomms15530 | Fab fragment was used for crystallization. See Materials and methods, ‘Crystallography’ subsection | |
| Recombinant DNA reagent | pAc-mfT1r3L-Ve | DOI:10.1038/srep25745 | ||
| Recombinant DNA reagent | pAc-mfT1r2aL-Ce | DOI:10.1038/srep25745 | ||
| Recombinant DNA reagent | pAc-mfT1r3L | DOI:10.1002/pro.3271 | ||
| Recombinant DNA reagent | pAc-mfT1r2aL | DOI:10.1002/pro.3271 | ||
| Sequence-based reagent | PCR primer used for T1r3-T105A mutation | This paper | TAC AAC GCG TGC AGA CAC TCA GCT GTT ATT G | |
| Sequence-based reagent | PCR primer used for T1r3-T105A mutation | This paper | TCT GCA CGC GTT GTA GAT TTT ATA ACC CAA C | |
| Peptide, recombinant protein | FLAG peptide | PH Japan | peptide | DYKDDDDK |
| Peptide, recombinant protein | gurmarin | DOI:10.1016/0300-9629(91)90,475r | peptide | Prof. Yuzo Ninomiya |
| Commercial assay or kit | Protein Thermal Shift Dye Kit | Thermo Fisher | Cat # 4461146 | |
| Software, algorithm | XDS | DOI:10.1107/S0907444909047337 | ||
| Software, algorithm | PHASER | DOI:10.1107/S0021889807021206 | ||
| Software, algorithm | Protein Thermal Shift Software | Applied Biosystems | Version 1.3 | |
| Software, algorithm | KaleidaGraph | Synergy Software | ||
| Software, algorithm | ORIGIN | OriginLab |





