Axon guidance genes modulate neurotoxicity of ALS-associated UBQLN2
Figures
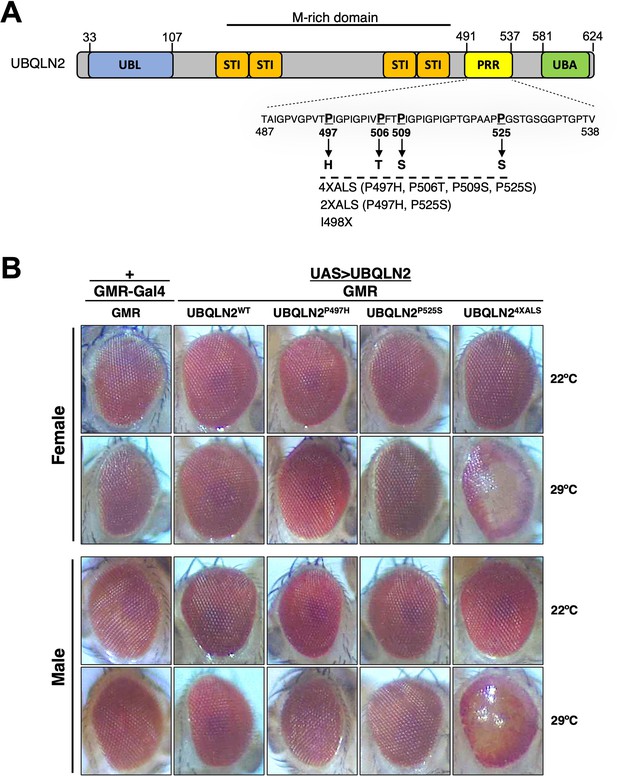
UBQLN2 exerts heat stress (HS)-dependent toxicity.
(A) Schematic of UBQLN2 and amyotrophic lateral sclerosis (ALS)-associated mutations. Approximate locations of ubiquitin-like (UBL); STI1-like (STI); proline-rich repeat (PRR); and ubiquitin-associated (UBA) domains are shown, as are ALS-associated mutations investigated in this study. (B) Eye images from flies expressing UBQLN2WT, UBQLN2P497H, UBQLN2P525S, or UBQLN24XALS under control of the eye-specific GMR driver at 22°C and 29°C. Note depigmentation and destruction of ommatidial facets in UBQLN24XALS flies reared at 29°C.
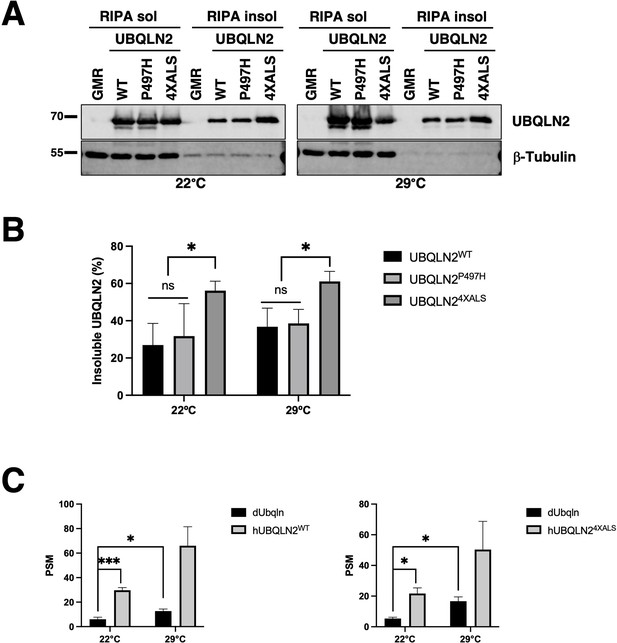
Ubiquitin (Ub)-binding contributes to eye degeneration and aggregation of UBQLN2ALS mutants.
(A) Expression and RIPA solubility of UBQLN2 proteins in head extracts prepared from GMR>UBQLN2 flies of the indicated genotype. Ten flies per genotype. (B) Quantification of RIPA solubility of UBQLN2 proteins in head extracts prepared from GMR>UBQLN2 flies of the indicated genotype. Triplicate of 10 flies per genotype. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. *p≤0.05, ***p≤0.001. (C) Relative abundance of Drosophila Ubqln (dUbqln) and human UBQLN2 (hUBQLN2) in GMR > UBQLN2 flies. Head extracts from flies of the indicated genotypes were analyzed by mass spectrometry (MS) to determine peptide spectral matches (PSM) for dUbqln and hUBQLN2 at 22°C and 29°C. MS was carried out using 100 flies per genotype. The bars represent mean with SEM from triplicate samples. Unpaired t-test was used for statistical analysis.
-
Figure 1—figure supplement 1—source data 1
Uncropped Western blot images corresponding to Figure 1—figure supplement 1A, and PSM corresponding to Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig1-figsupp1-data1-v2.zip
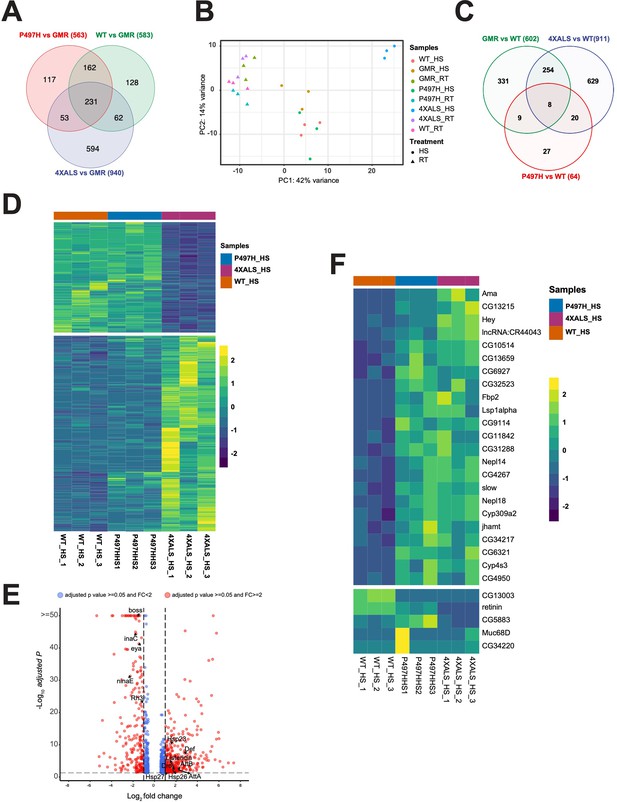
Gene expression profiles of GMR>UBQLN2ALS flies.
(A) Venn diagram of differentially expressed genes (DEGs) (amyotrophic lateral sclerosis [ALS]) from comparisons of GMR>UBQLN2WT, GMR>UBQLN2P497H, and GMR>UBQLN24XALS to GMR-Gal4 control flies reared at 29°C. (B) Principle component analysis of RNA-Seq data from whole heads of GMR, GMR>UBQLN2WT, GMR>UBQLN2P497H, and GMR>UBQLN24XALS flies reared at 22°C or 29°C. (C) Venn diagram of DEGs from comparisons of GMR-Gal4 control, GMR>UBQLN2P497H, and GMR>UBQLN24XALS to GMR>UBQLN2WTflies reared at 29°C. (D) Heat map of genes differentially expressed between GMR>UBQLN2WT and GMR>UBQLN24XALS fly heads at 29°C. (E) Volcano plot depicting genes differentially expressed between GMR>UBQLN2WT and GMR>UBQLN24XALS flies at 29°C. Downregulated photoreceptor genes, upregulated innate immunity genes, and upregulated small HSPs are highlighted. (F) DEGs common to GMR>UBQLN2P497H and GMR>UBQLN24XALS flies in comparison to GMR>UBQLN2WT flies at 29°C.
-
Figure 1—figure supplement 2—source data 1
Gene expression data sets corresponding to Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig1-figsupp2-data1-v2.zip
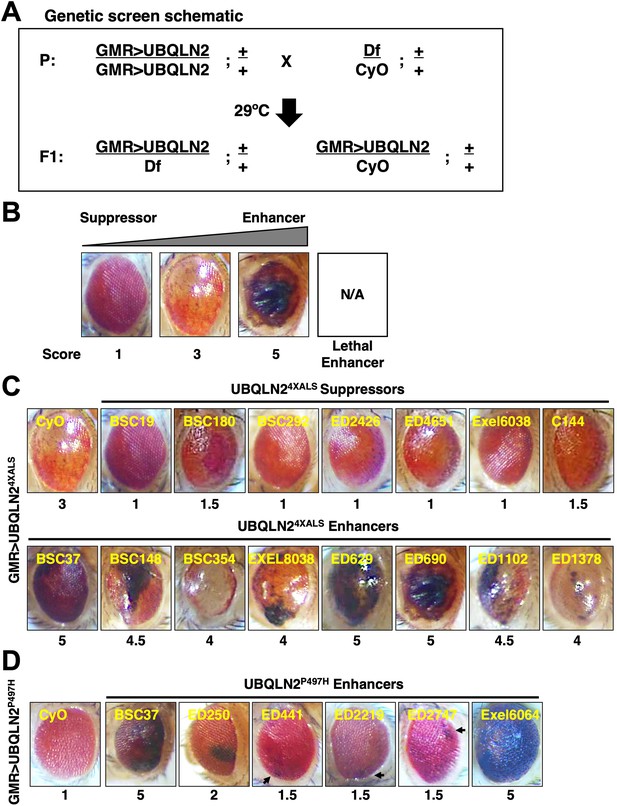
Identification of UBQLN2 modifier genes.
(A) Schematic of deficiency (Df) screen for UBQLN2 modifiers. (B) Representative eye images and scoring rubric for F1 progeny of GMR>UBQLN24XALS flies crossed to Df lines. (C) Representative UBQLN24XALS modifier genes. Eye images were taken of F1 progeny from crosses of GMR>UBQLN24XALS to indicated Df lines at 1–3 days post eclosion. Suppressors and enhancers are shown in top and bottom rows, respectively. (D) Representative eye images of UBQLN2P497H enhancers. Arrows indicate foci of eye degeneration. Eye degeneration scores are displayed below each eye image (B, C, D).
-
Figure 2—source data 1
List of Dfs that modified GMR>UBQLN2P497H and/or GMR>UBQLN24XALS eye phenotypes corresponding to Figure 2C, D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig2-data1-v2.zip
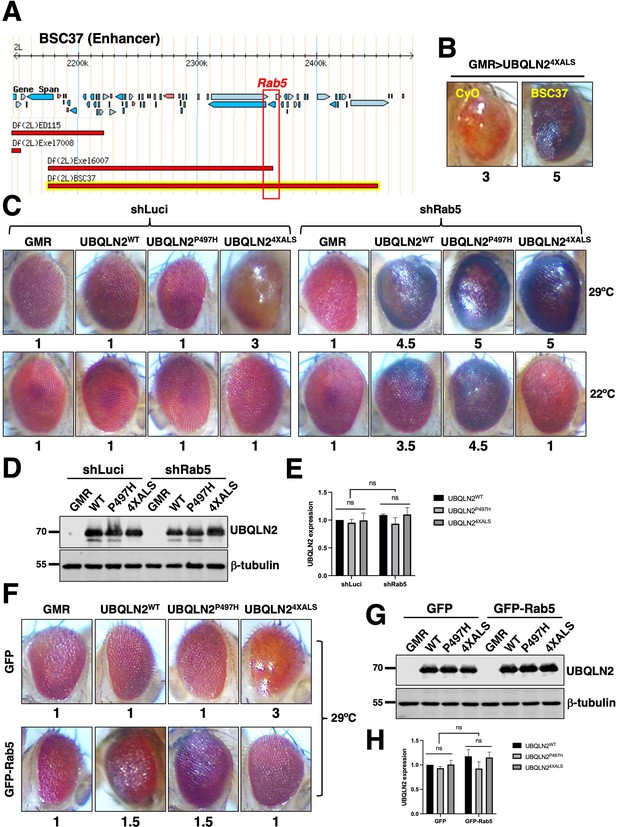
Rab5 is a UBQLN24XALS modifier gene.
(A) Genomic map of BSC37. The region spanning the Rab5 gene is boxed. (B) Representative eye images from GMR>UBQLN24XALS/BSC37 and GMR>UBQLN24XALS/CyO flies. (C) Differential effects of Rab5 knockdown on GMR>UBQLN2 eye phenotypes at 22°C and 29°C. Recombinant GMR>UBQLN2 flies of indicated genotypes were crossed to flies expressing shRNAs targeting Rab5 (shRab5) or luciferase (shLuci). F1 progeny were processed for eye imaging 2–3 days after eclosion. (D, E) UBQLN2 expression levels in whole heads of GMR-Gal4, GMR>UBQLN2WT, GMR>UBQLN2P497H, or GMR>UBQLN24XALS flies on the genetic backgrounds of shLuci or shRab5. (E) Rab5 knockdown did not impact UBQLN2 expression. Quantification of UBQLN2 expression normalized to β-tubulin. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. (F) GFP-Rab5 overexpression rescued the UBQLN24XALS RE phenotype at 29°C. GMR>UBQLN2 flies of indicated genotypes were crossed to flies harboring UAS-GFP or UAS-GFP-Rab5 transgenes. Eye degeneration scores are displayed below each eye image (B, C, F). (G, H) UBQLN2 expression levels in whole heads of GMR-Gal4, GMR>UBQLN2WT, GMR>UBQLN2P497H, or GMR>UBQLN24XALS flies on the genetic backgrounds of GFP or GFP-Rab5 overexpression. (H) Quantification of UBQLN2 expression normalized to β-tubulin. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis.
-
Figure 2—figure supplement 1—source data 1
Uncropped Western blot images corresponding to Figure 2—figure supplement 1D, E, G, H.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig2-figsupp1-data1-v2.zip
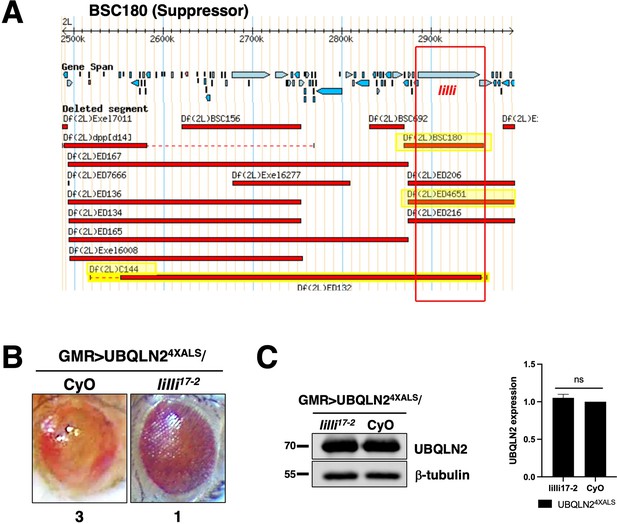
Transcriptional elongation factor lilliputian (lilli) is a UBQLN24XALS suppressor.
(A) Schematic of BSC180, ED4651, C144, and the lilli gene locus. (B) Reduced expression of lilli rescued the UBQLN24XALS RE phenotype at 29°C. Representative eye phenotypes from GMR>UBQLN24XALS flies harboring the indicated alleles. Eye degeneration scores are displayed below each eye image. (C) UBQLN2 expression levels in whole heads of GMR>UBQLN24XALS flies on the genetic backgrounds of lilli mutation or CyO control. Quantification of UBQLN2 expression normalized to β-tubulin (right panel). The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis.
-
Figure 2—figure supplement 2—source data 1
Uncropped Western blot images corresponding to Figure 2—figure supplement 2C.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig2-figsupp2-data1-v2.zip
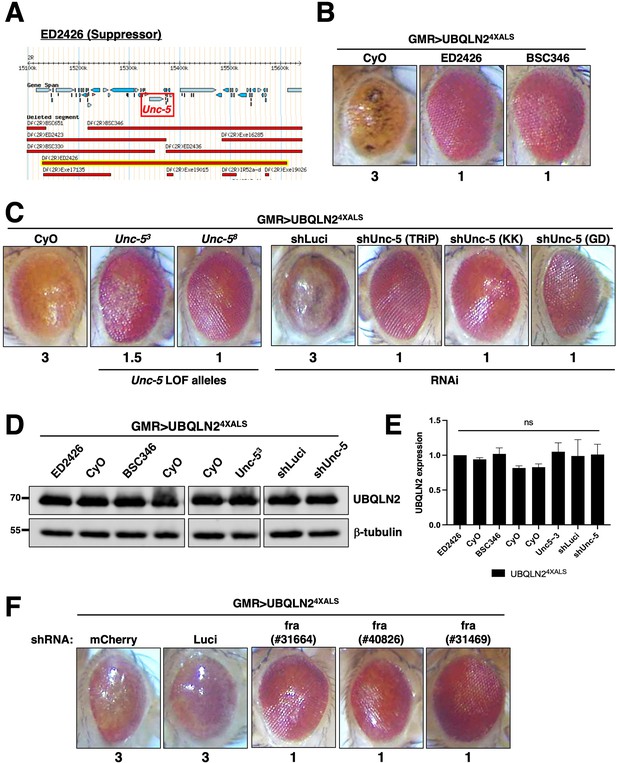
Disruption of Unc-5 suppresses UBQLN2-associated eye degeneration.
(A) Schematic depiction of ED2426 and overlapping deficiencies (Dfs) in relation to the Unc-5 gene locus. (B) Representative eye images of GMR>UBQLN24XALS flies crossed to Df lines ED2426 and BSC346. (C) Single allele expression of Unc-5 LOF alleles (left panels) or three independent Unc-5 RNAi alleles diminished the rough eye (RE) phenotype of GMR>UBQLN24XALS flies at 29°C. (D, E) UBQLN2 expression levels in whole heads of GMR>UBQLN24XALS flies on the indicated genetic backgrounds. (E) Quantification of UBQLN2 expression normalized to β-tubulin. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. (F) fra silencing reduced the RE phenotype of GMR>UBQLN24XALS flies. Shown are representative eye phenotypes of F1 progeny from GMR>UBQLN24XALS flies crossed to control (shLuci, shmCherry), or fra RNAi lines at 29°C. Eye degeneration scores are displayed below each eye image (B, C, F).
-
Figure 3—source data 1
Uncropped Western blot images corresponding to Figure 3D, E.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig3-data1-v2.zip
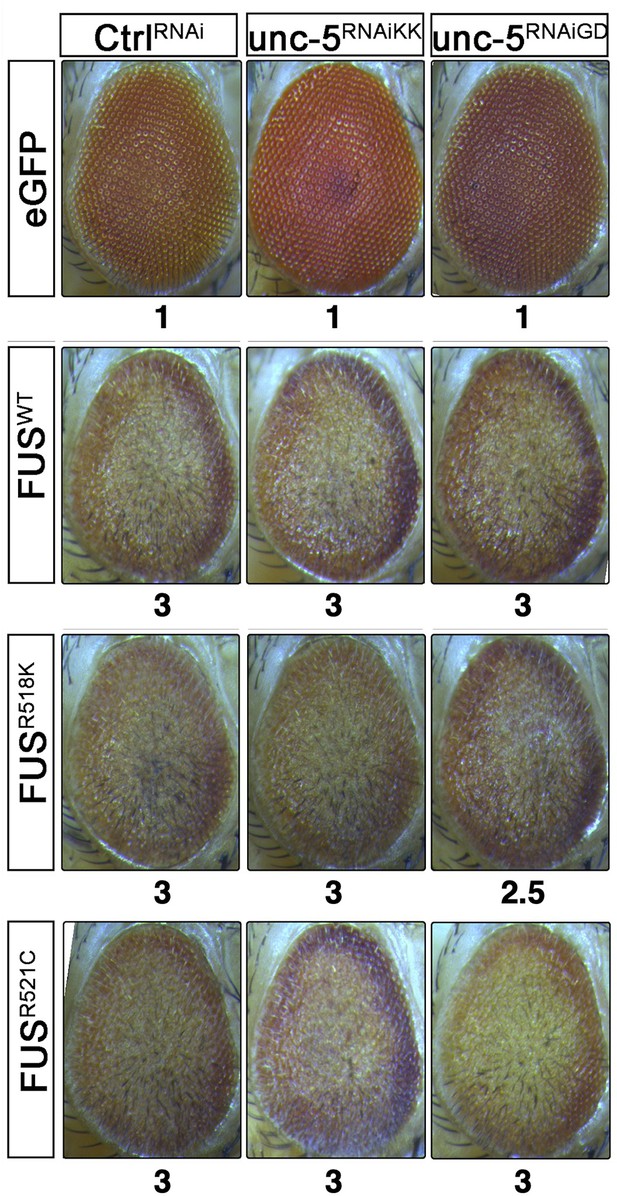
Unc-5 silencing does not modify FUS-associated eye degeneration.
Representative eye images of GMR>eGFP (control), GMR>FUSWT, GMR>FUSR518K, or GMR>FUSR521C flies harboring the indicated shRNA alleles. Eye degeneration scores are displayed below each eye image.
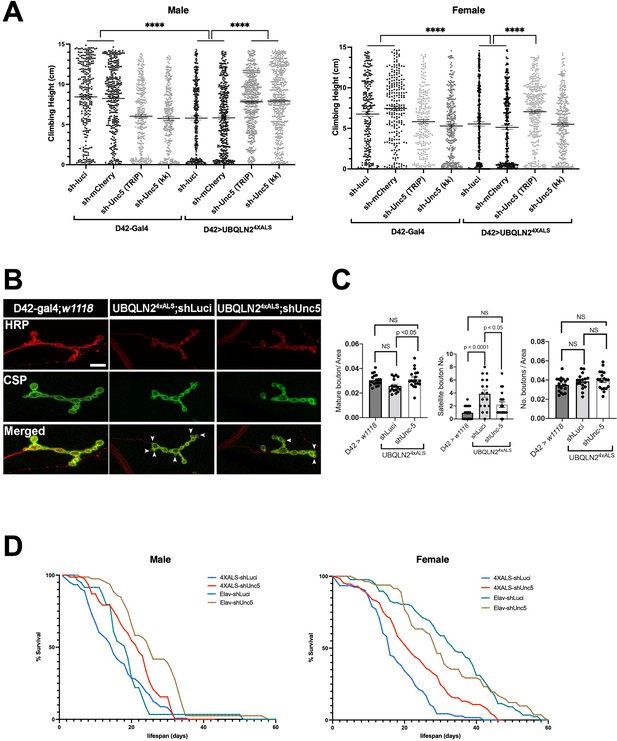
Unc-5 silencing in Drosophila suppressed UBQLN24XALS-associated neuronal phenotypes.
(A) Recombinant D42>UBQLN24XALS flies expressing UBQLN2 under control of the motor neuron-specific D42 driver were crossed to the indicated RNAi lines. Climbing potential of male (left) and female (right) progeny reared at 29°C was measured 7 days after eclosion as described in Materials and methods. Data analysis was performed using ordinary one-way ANOVA. Data are shown as mean ± SEM. n>100 flies, ****p≤0.0001. (B) Neuromuscular junction (NMJ) morphology analysis of D42>UBQLN24XALS larvae expressing the indicated shRNAs. NMJs dissected from third instar larvae were stained with α-HRP and α-CSP antibodies and imaged by confocal microscopy. Scale bar: 10μm. (C) Number of NMJs harboring indicated phenotypes were tabulated from greater than 50 NMJs per genotype. Unpaired t-test was used for statistical analysis. Data are shown as mean ± SEM. (D) Pan-neuronal Unc-5 knockdown enhances lifespan of Elav>UBQLN24XALS flies. Recombinant Elav>UBQLN24XALS or a Elav>Gal4 flies were crossed to the indicated RNAi lines (shLuci or shUnc5). Lifespan of male (left panel) and female (right panel) progeny reared at 27°C was measured as described in Materials and methods. n>50 flies.
-
Figure 4—source data 1
Data sets corresponding to Figure 4A, C, D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig4-data1-v2.zip
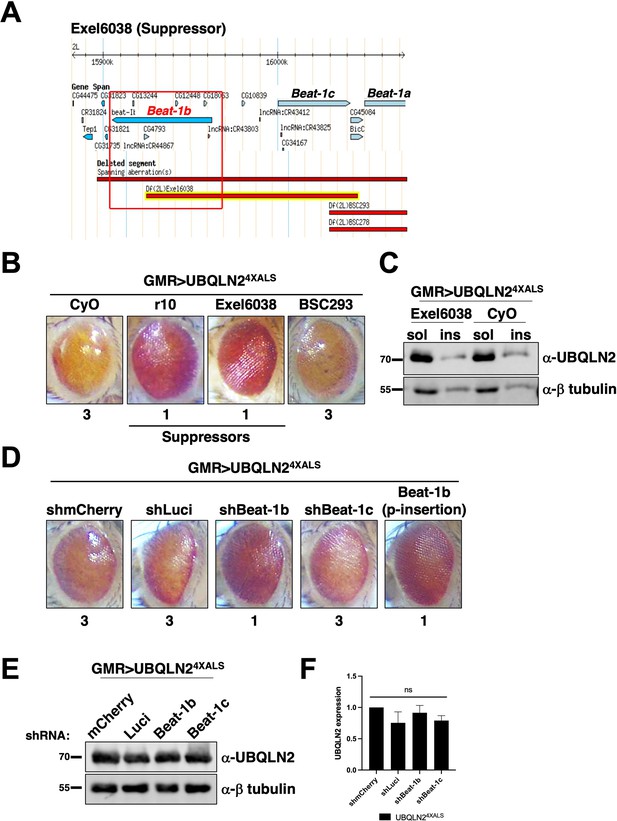
The axon guidance gene beat-1b is a UBQLN24XALS suppressor.
(A) Schematic of the beat-1b gene locus. (B) Representative eye phenotypes of GMR>UBQLN24XALS flies harboring the indicated deficiency (Df) alleles. (C) UBQLN2 expression levels and RIPA solubility in whole heads of GMR>UBQLN24XALS/Exel6038 and GMR>UBQLN24XALS/CyO flies. (D) Representative eye phenotypes of GMR>UBQLN24XALS flies expressing the indicated shRNAs. Eye degeneration scores are displayed below each eye image (B, D). (E, F) Knockdown of Beat-1b or Beat-1c does not inhibit UBQLN2 expression in GMR>UBQLN24XALS flies. (F) Quantification of UBQLN2 expression normalized to β-tubulin. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. .
-
Figure 5—source data 1
Uncropped Western blot images corresponding to Figure 5C, E, F.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig5-data1-v2.zip
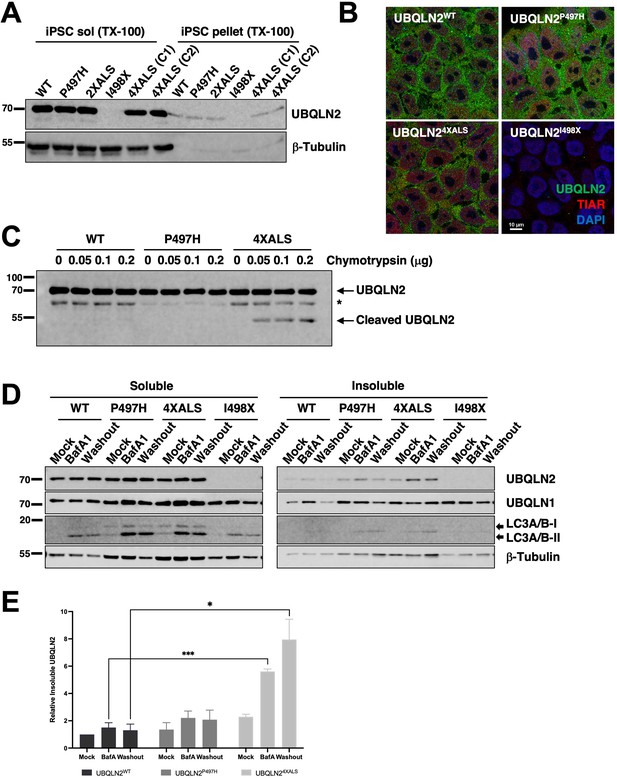
Localization and solubility of UBQLN2ALS mutants in induced pluripotent stem cells (iPSCs).
(A) Extracts from UBQLN2WT, UBQLN2P497H, UBQLN22XALS, UBQLN2I498X, or UBQLN24XALS iPSCs (clone 1 [C1] and clone 2 [C2]) were separated into soluble and insoluble fractions in 1% Triton X-100 (TX-100) buffer and immunoblotted with α-UBQLN2, and α-β-tubulin antibodies. (B) Localization patterns of wild-type and UBQLN2ALS proteins in iPSCs. UBQLN2WT, UBQLN2P497H, UBQLN24XALS, and UBQLN2I498X iPSCs were stained with α-UBQLN2 and α-TIAR antibodies and imaged by confocal microscopy. Note the lack of cytosolic aggregates. (C) Cell extracts from iPSCs of the indicated genotypes were incubated at room temperature with increasing amounts of chymotrypsin for 5 min. After separation by SDS-PAGE, the proteins were immunoblotted with α-UBQLN2 antibodies. Positions of full-length and cleaved UBQLN2 are denoted by arrows. *: non-specific band. (D) Autophagy inhibition with BafA1 reduced solubility of endogenous UBQLN2ALS proteins. UBQLN2WT, UBQLN2P497H, UBQLN24XALS, and UBQLN2I498X iPSCs were treated with 100 nM of BafA1 for 16 hr followed by BafA1 washout and incubation in BafA1-free growth media for 8 hr. Detergent extracts were separated into soluble and insoluble fractions and analyzed by SDS-PAGE and immunoblotting using UBQLN2, UBQLN1, LC3A/B, and β-tubulin antibodies. (E) Quantification of UBQLN2 solubility in UBQLN2WT, UBQLN2P497H, and UBQLN24XALS iPSCs from (D). The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. *p≤0.05, ***p≤0.001.
-
Figure 6—source data 1
Uncropped Western blot images corresponding to Figure 6A, C, D, E.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig6-data1-v2.zip
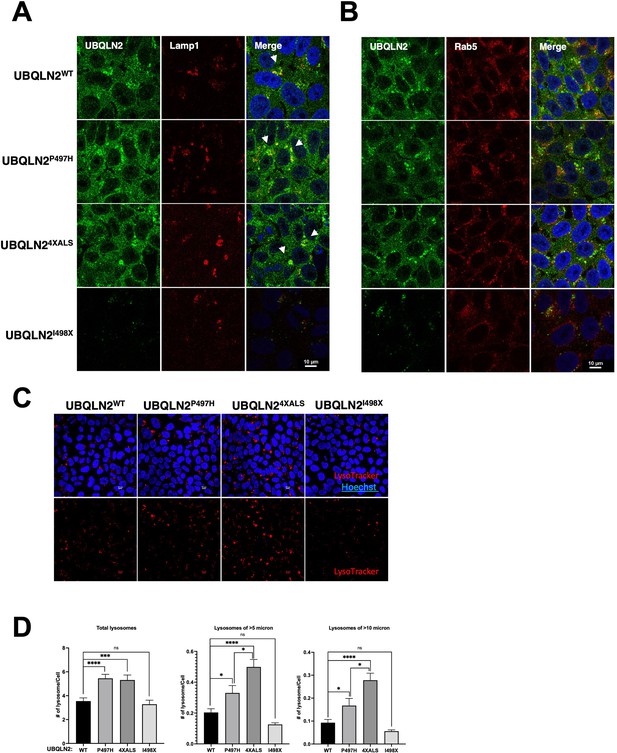
Endogenous UBQLN2ALS mutants perturb lysosomes.
(A) UBQLN2WT, UBQLN2P497H, UBQLN24XALS, and UBQLN2I498X induced pluripotent stem cells (iPSCs) were treated with 100 nM of BafA1 for 16 hr and processed for immunostaining with UBQLN2 and Lamp1 antibodies. Arrowheads indicate colocalization of UBQLN2 and Lamp1. (B) UBQLN2WT, UBQLN2P497H, UBQLN24XALS, and UBQLN2I498X iPSCs were immunostained with UBQLN2 and Rab5 antibodies after incubation in 100 nM BafA1 for 16 hr. (C, D) Increased lysosomal size and number in UBQLN2ALS mutant iPSCs. (C) Representative images from UBQLN2WT, UBQLN2P497H, UBQLN24XALS, and UBQLN2 I498X iPSCs following labeling with LysoTracker Red DND-99 for 1 hr and costaining with Hoechst 33342. Scale bar: 10μm. (D) Size and numbers of LysoTracker-positive compartments were analyzed on a per cell basis using Fiji. Unpaired t-test was used for statistical analysis. Data are shown as mean ± SEM. *p≤0.05, ***p≤0.001, ****p≤0.0001.
-
Figure 6—figure supplement 1—source data 1
Data sets corresponding to Figure 6—figure supplement 1C, D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig6-figsupp1-data1-v2.zip
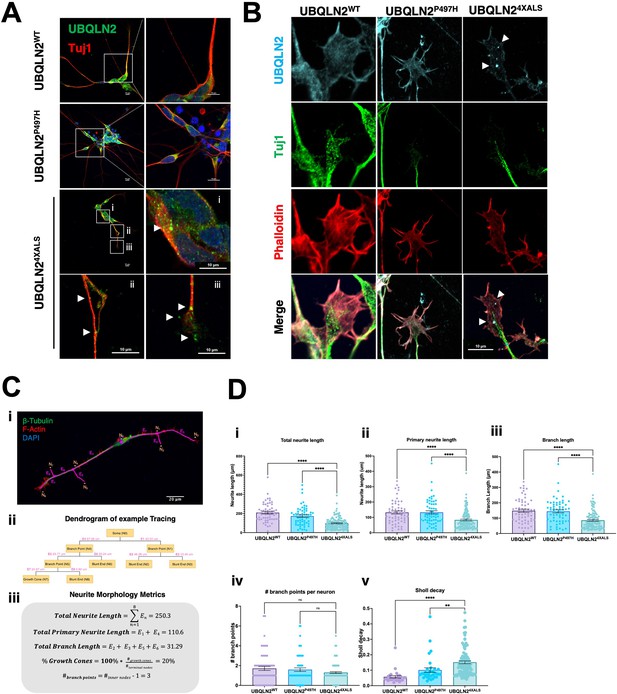
Protein aggregation and neurite defects in UBQLN2ALS inducible motor neurons (iMNs).
(A) UBQLN2WT, UBQLN2P497H, and UBQLN24XALS iMNs were immunostained for UBQLN2 and Tuj1. Magnified images of UBQLN24XALS iMN soma (i), axons (ii), and neurite terminals (iii) are shown; aggregates are marked with arrows. (B) UBQLN2 localizes to growth cone lamellipodia and filopodia. Differentiated iMNs of the indicated genotypes were costained for UBQLN2, Tuj1, and filamentous actin (phalloidin). Note reduced complexity of the UBQLN24XALS growth cone. Arrowheads indicate UBQLN24XALS aggregates. Scale bars = 10 μm. (C) Schematic of Sholl analysis. Tracing example of an iMN (i) displays the paths representing individual neuron structure, with nodes (Ni) and edges (Ej) corresponding to the dendrogram on (ii) and (iii). (D) UBQLN24XALS iMNs exhibit reduced complexity. UBQLN2WT, UBQLN2P497H, and UBQLN24XALS iMNs were stained with α-Tuj1 and imaged by confocal microscopy. One hundred neurons of the indicated genotypes were traced using Simple Neurite Tracer (SNT) and subjected to Sholl image analysis to quantify total neurite projection path length (i), primary neurite length (ii), terminal neurite length (iii), neurite branch points (iv), and Sholl decay (v). Data analysis was performed using ordinary one-way ANOVA. Data are shown as mean ± SEM. n>100 iMNs, **p≤0.01, ****p≤0.0001.
-
Figure 7—source data 1
Data sets corresponding to Figure 7D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig7-data1-v2.zip
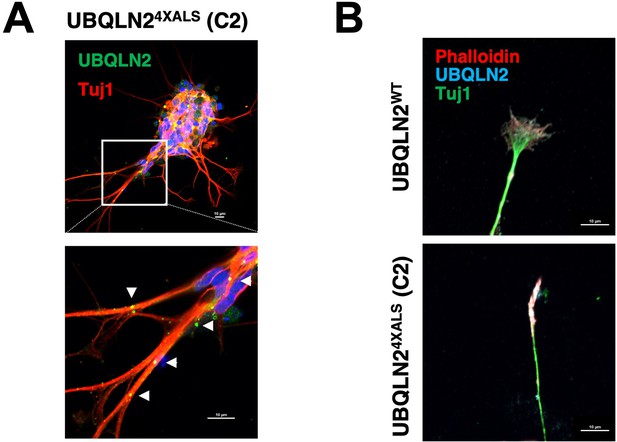
Protein aggregation and reduced neurite complexity in an independent UBQLN24XALS inducible motor neuron (iMN) line.
(A) UBQLN24XALS (C2) iMNs were immunostained for UBQLN2 and Tuj1. Aggregates are marked with arrowheads. (B) Growth cone morphologies in UBQLN24XALS (C2) iMNs. iMNs were stained with α-UBQLN2, α-Tuj1, and phalloidin. Note reduced growth cone elaboration in UBQLN24XALS (C2) iMNs. Scale bars = 10 μm.
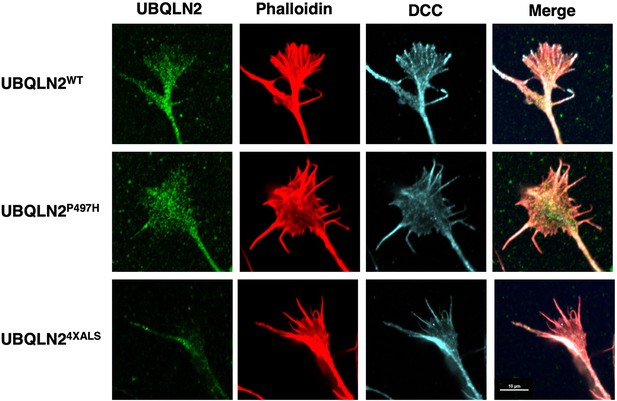
The UBQLN24XALS mutation did not significantly impact the localization of DCC.
Inducible motor neurons (iMNs) of the indicated genotypes were immunostained with α-UBQLN2 and α-DCC antibodies, and phalloidin. Scale bars = 10 μm.
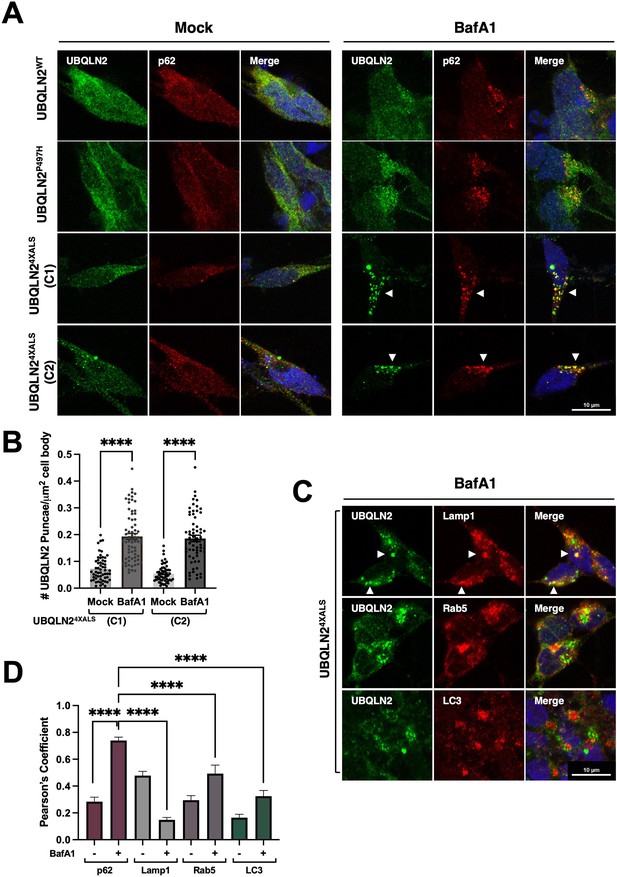
BafA1 induces p62-positive UBQLN24XALS aggresomes in inducible motor neurons (iMNs).
(A) UBQLN2WT, UBQLN2P497H, and UBQLN24XALS iMNs were treated with BafA1 (50 nM) for 16 hr and processed for immunostaining with UBQLN2 and p62. Arrowheads indicate UBQLN24XALS aggregates that were colocalized with p62. (B) The number of UBQLN24XALS aggregates was analyzed on a per cell basis using Fiji. Data analysis was performed using ordinary one-way ANOVA. Data are shown as mean ± SEM. n>50 iMNs, ****p≤0.0001. (C) UBQLN24XALS iMNs were treated with BafA1 and processed for immunostaining with UBQLN2 and Lamp1, Rab5, or LC3. Arrowheads indicate UBQLN24XALS colocalization with Lamp1. Scale bars = 10 μm. (D) Pearson’s correlation coefficients for colocalization assays. Pearson’s coefficients were plotted as a bar graph to compare the colocalization of UBQLN2 with p62, LAMP1, Rab5, or LC3. n=10 iMNs, Error bars represent SEM, ****p≤0.0001 (ordinary one-way ANOVA).
-
Figure 8—source data 1
Data sets corresponding to Figure 8B, D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig8-data1-v2.zip
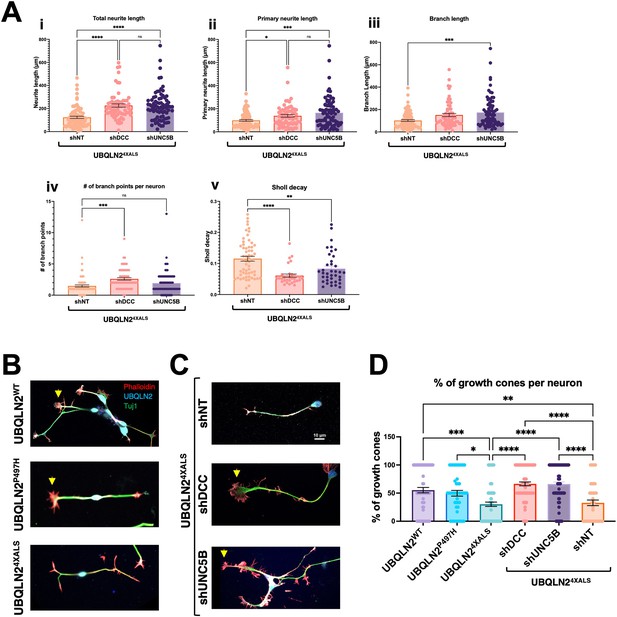
UNC5B and DCC silencing partially reverse neurite and growth cone defects in UBQLN24XALS inducible motor neurons (iMNs).
(A) UBQLN24XALS iMNs expressing shNT, shUNC5B, or shDCC were subjected to Sholl analysis of neurite length and complexity as described in Figure 7. Data analysis was performed using ordinary one-way ANOVA. Data are shown as mean ± SEM. n>100 iMNs, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. (B) Growth cone morphologies in UBQLN24XALS iMNs. UBQLN2WT, UBQLN2P497H, and UBQLN24XALS iMNs. iMNs of the indicated genotypes were stained with α-UBQLN2, α-Tuj1, and phalloidin. Note reduced growth cone elaboration in UBQLN24XALS iMNs. (C) Enhanced growth cone elaboration in UBQLN24XALS iMNs expressing DCC or UNC5B shRNAs. iMNs of the indicated genotype were stained with α-UBQLN2, α-Tuj1, and phalloidin. Arrowheads denote growth cones. (D) Quantification of growth cones in UBQLN2WT, UBQLN2P497H, and UBQLN24XALS, and UBQLN24XALS iMNs expressing the indicated shRNAs. Data analysis was performed using ordinary one-way ANOVA. Data are shown as mean ± SEM. n>100 iMNs, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
-
Figure 9—source data 1
Data sets corresponding to Figure 9A, D.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig9-data1-v2.zip
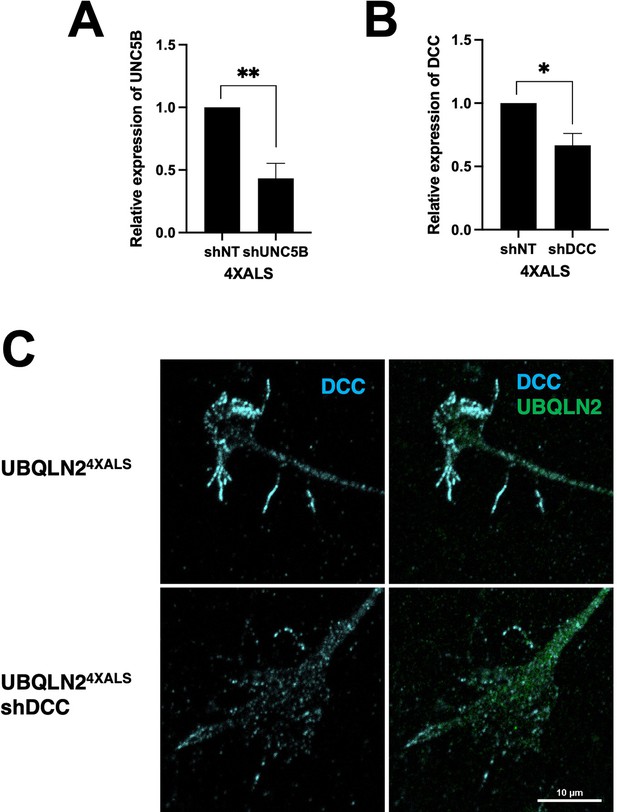
Expression of UNC5B and DCC mRNA in UBQLN24XALS induced pluripotent stem cells (iPSCs) transduced with lentiviral shRNA vectors.
mRNA levels of UNC5B (A) and DCC (B) were analyzed by RT-qPCR in iPSCs expressing the indicated shRNAs. The bars represent mean with SEM of triplicate samples. Unpaired t-test was used for statistical analysis. *p≤0.05, **p≤0.01. (C) Inducible motor neurons (iMNs) of the indicated genotypes were immunostained with α-DCC antibodies. Note reduced filopodial DCC signal intensity in UBQLN24XALS: shDCC iMNs.
-
Figure 9—figure supplement 1—source data 1
Data sets corresponding to Figure 9—figure supplement 1A, B.
- https://cdn.elifesciences.org/articles/84382/elife-84382-fig9-figsupp1-data1-v2.zip
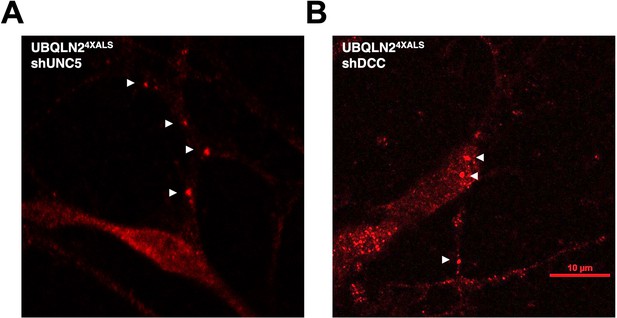
UNC5B or DCC knockdown does not affect UBQLN24XALS aggregation in inducible motor neurons (iMNs).
UNC5B (A) or DCC (B) knockdown iMNs were immunostained with α-UBQLN2 antibodies. Aggregates are marked with arrowheads.
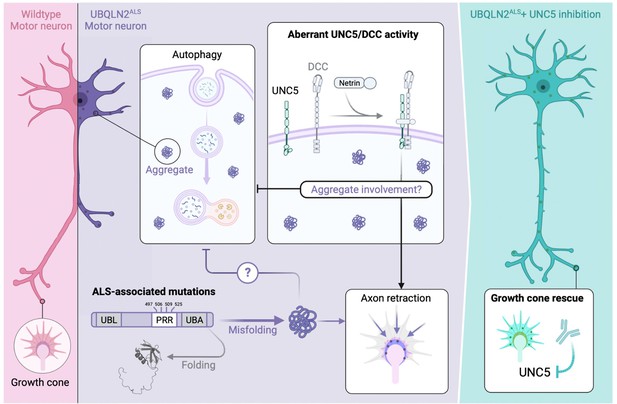
Speculative model for UBQLN2ALS toxicity suppression through the UNC5 pathway.
(Left panel) Wild-type motor neurons exhibit healthy neurites and well-elaborated growth cones devoid of UBQLN2 aggregates. (Center panel) Amyotrophic lateral sclerosis (ALS)-associated mutations in the proline-rich repeat (PRR) of UBQLN2 promote its misfolding and assembly into aggregates that trigger UNC5/DCC-dependent axonal retraction. UBQLN2ALS aggregates may directly initiate pathologic UNC5/DCC signaling or impact other aspects of cellular regulation, including the endolysosomal-autophagosomal pathway, leading to proteostasis deregulation and growth cone retraction. (Right panel) Suppression of UNC5/DCC signaling with rationally designed antibodies or small molecules (not shown) rescues growth cone defects in UBQLN2ALS motor neurons.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | Deficiency | BDSC | BDSC7521 | w[1118]; Df(2L)Exel6038, P{w[+mC]=XPU}Exel6038/CyO |
| Genetic reagent (D. melanogaster) | beat-1c/RNAi | BDSC | BDSC64528 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMC05547}attP40 |
| Genetic reagent (D. melanogaster) | beat-1b/RNAi | BDSC | BDSC55938 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMC04226}attP40 |
| Genetic reagent (D. melanogaster) | Rab5/RNAi | BDSC | BDSC34832 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMS00147}attP2 |
| Genetic reagent (D. melanogaster) | Luciferase/RNAi | BDSC | BDSC31603 | y(1) v(1); P{y[+t7.7] v[+t1.8]=TRiP.JF01355}attP2 |
| Genetic reagent (D. melanogaster) | Rab5/RNAi | BDSC | BDSC30518 | y(1) v(1); P{y[+t7.7] v[+t1.8]=TRiP.JF03335}attP2 |
| Genetic reagent (D. melanogaster) | beat-1b/P element | BDSC | BDSC18802 | w[1118]; PBac{w[+mC]=WH}beat-Ib[f04746] |
| Genetic reagent (D. melanogaster) | Rab5/overexpression | BDSC | BDSC43336 | w[*]; P{w[+mC]=UAS-GFP-Rab5}3 |
| Genetic reagent (D. melanogaster) | Unc-5/RNAi | BDSC | BDSC33756 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMS01099}attP2 |
| Genetic reagent (D. melanogaster) | Unc-5/RNAi | VDRC | VDRC8138 | GD RNAi |
| Genetic reagent (D. melanogaster) | Unc-5/RNAi | VDRC | VDRC110155 | KK RNAi |
| Genetic reagent (D. melanogaster) | GFP/overexpression | BDSC | BDSC5430 | w[1118]; P{w[+mC]=UAS-EGFP}34/TM3, Sb(1) |
| Genetic reagent (D. melanogaster) | Deficiency | BDSC | BDSC24370 | w[1118]; Df(2R)BSC346/CyO |
| Genetic reagent (D. melanogaster) | mCherry/RNAi | BDSC | BDSC35787 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=UAS-mCherry.VALIUM10}attP2 |
| Genetic reagent (D. melanogaster) | frazzled/RNAi | BDSC | BDSC31469 | y(1) v(1); P{y[+t7.7] v[+t1.8]=TRiP.JF01231}attP2 |
| Genetic reagent (D. melanogaster) | frazzled/RNAi | BDSC | BDSC31664 | y(1) v(1); P{y[+t7.7] v[+t1.8]=TRiP.JF01457}attP2 |
| Genetic reagent (D. melanogaster) | frazzled/RNAi | BDSC | BDSC40826 | y(1) sc[*] v(1) sev(21); P{y[+t7.7] v[+t1.8]=TRiP.HMS01147}attP2 |
| Genetic reagent (D. melanogaster) | Deficiency (lilli) | BDSC | BDSC9610 | BSC180 |
| Genetic reagent (D. melanogaster) | Deficiency (Rab5) | BDSC | BDSC7144 | BSC37 |
| Genetic reagent (D. melanogaster) | Deficiency (lilli) | BDSC | BDSC94697 | ED4651 |
| Genetic reagent (D. melanogaster) | Deficiency (lilli) | BDSC | BDSC99 | C144 |
| Genetic reagent (D. melanogaster) | Lilli (LOF allele) | BDSC | BDSC5726 | lilli[A17-2] cn(1) bw(1)/CyO |
| Genetic reagent (D. melanogaster) | Unc-5 (LOF) | Greg Bashaw; Labrador et al., 2005 | https://doi.org/10.1016/j.cub.2005.06.058 | Unc-53 |
| Genetic reagent (D. melanogaster) | Unc-5 (LOF) | Greg Bashaw; Labrador et al., 2005 | https://doi.org/10.1016/j.cub.2005.06.058 | Unc-58 |
| Genetic reagent (D. melanogaster) | Unc-5/overexpression | Greg Bashaw | HA-Unc-5 (Chr2) | |
| Genetic reagent (D. melanogaster) | Unc-5/overexpression | Greg Bashaw | HA-Unc-5 (Chr3) | |
| Cell line(Homo sapiens) | UBQLN2WT | WC031i-5907–6 | WT | |
| Cell line (Homo sapiens) | UBQLN2P497H | This paper | P497H | Mutation using CRISPR |
| Cell line (Homo sapiens) | UBQLN22XALS | This paper | P497H, P525S | Mutation using CRISPR |
| Cell line (Homo sapiens) | UBQLN24XALS | This paper | P497H, P506T, P509S, P525S | Mutation using CRISPR |
| Cell line (Homo sapiens) | UBQLN2I498X | This paper | I498X | Mutation using CRISPR |
| Transfected construct (humani PSCs) | shUNC5B | Sigma-Aldrich | TRCN0000442978 | Lentiviral construct to transfect and express the shRNA in iPSCs |
| Transfected construct (human iPSCs) | shDCC | Sigma-Aldrich | TRCN0000010318 | Lentiviral construct to transfect and express the shRNA in iPSCs |
| Antibody | Anti-UBQLN2 (Mouse monoclonal antibody) | Abcam | Cat#: Ab190283 | IF (1:1000), WB (1:10000) |
| Antibody | Anti-UBQLN2 (Rabbit polyclonal antibody) | Cell Signaling Technology | Cat#: 85509 | IF (1:500), WB (1:2000) |
| Antibody | Anti-β-Tubulin (Mouse monoclonal antibody) | EMD Millipore | Cat#: 05–661 | WB (1:2000) |
| Antibody | Anti-LC3A/B (Rabbit polyclonal antibody) | Cell Signaling Technology | Cat#: 12741S | IF (1:500), WB (1:1000) |
| Antibody | Anti-LAMP1 (Mouse monoclonal antibody) | Santa Cruz Biotechnology | Cat#: sc-20011 | IF (1:500), WB (1:1000) |
| Antibody | Anti-Rab5A (Rabbit polyclonal antibody) | Cell Signaling Technology | Cat#: 46449S | IF (1:500), WB (1:1000) |
| Antibody | Anti-UBQLN1 (Rabbit polyclonal antibody) | Cell Signaling Technology | Cat#: 14526 | IF (1:500), WB (1:2000) |
| Antibody | Anti-Tuj1 (Mouse monoclonal antibody) | EMD Millipore | Cat#: MAB1637MI | IF (1:500) |
| Antibody | Anti-DCC (Rabbit polyclonal antibody) | Invitrogen | Cat#: PA5-50946 | IF (1:500) |
| Antibody | Anti-DLG (Mouse monoclonal antibody) | DSHB | Cat#: 4F3 | IF (1:100) |
| Antibody | Anti-HRP- Cy3-conjugated (Goat polyclonal antibody) | Jackson ImmunoResearch | Cat#:123-165-021 | IF (1:100) |
| Sequence-based reagent | GAPDH-F | Sigma-Aldrich | qPCR primers | GTCTCCTCTGACTTCAACAGCG |
| Sequence-based reagent | GAPDH-R | Sigma-Aldrich | qPCR primers | ACCACCCTGTTGCTGTAGCCAA |
| Sequence-based reagent | UNC5B-F | Sigma-Aldrich | pPCR primers | ACTGCCGTGACTTCGACAC |
| Sequence-based reagent | UNC5B-R | Sigma-Aldrich | qPCR primers | GCCTTGCCGTCTTAAAGTTGA |
| Sequence-based reagent | DCC-F | Sigma-Aldrich | qPCR primers | GACTTTACCAATGTGAGGCATCT |
| Sequence-based reagent | DCC-R | Sigma-Aldrich | qPCR primers | GGTCCTGCTACTGCAACTTTT |
| Chemical compound, drug | CHIR99021 | Tocris | 4423 | Chemical compound, drug |
| Chemical compound, drug | DMH-1 | Tocris | 4126 | Chemical compound, drug |
| Chemical compound, drug | SB431542 | Stemgent | 04-0010 | Chemical compound, drug |
| Chemical compound, drug | Retinoic acid | Stemgent | 04-0021 | Chemical compound, drug |
| Chemical compound, drug | Purmorphamine | Stemgent | 04-0009 | Chemical compound, drug |
| Chemical compound, drug | Compound E | EMD Millipore | 565790 | Chemical compound, drug |
| Software, algorithm for RNA-Seq analysis | Drosophila melanogaster genome (dmel-all-chromosome-r6.27, FlyBase) | https://github.com/ENCODE-DCC/rna-seq-pipeline; ENCODE DCC, 2022 STAR 2.7.1a | DESeq2, MetaScape website | |
| Software, algorithm for mass spectrometry analysis | MetaMorpheus software program | FlashLFQ |





