Structural foundation for the role of enterococcal PrgB in conjugation, biofilm formation, and virulence
Figures
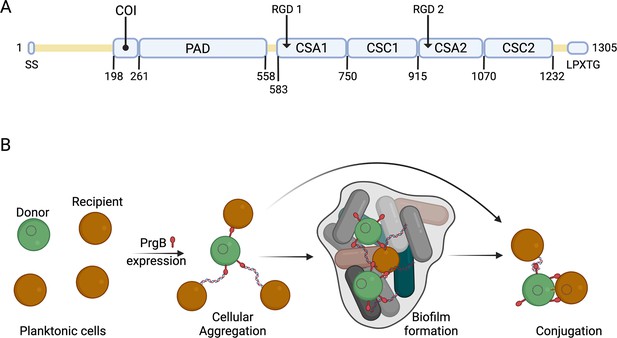
Schematic overview of PrgB domain organization and function.
(A) Updated schematic overview of the domain organization of PrgB. SS: signal sequence, COI: coiled-coil, PAD: polymer adhesin domain, CSA: adhesin isopeptide-forming adherence domain, CSC: cell-surface antigen C-terminal domain, LPXTG: cell wall anchor sequence. The PrgA cleavage site is located between the polymer adhesin domain and the first immunoglobulin (Ig)-like domain, and has the sequence IFNYGNPKEP. (B) In a setting with a donor cell (green) and multiple recipient cells (brown), PrgB is produced and sits on the cell wall. There it enhances cellular aggregation and/or biofilm formation, either by directly binding lipoteichoic acid (LTA) from the cell wall of a recipient or by binding first to extracellular DNA (eDNA). PrgB compacts the eDNA, and thereby likely pulls the recipient cells closer. Once close enough, PrgB binds to the LTA of the recipient bacteria and facilitates mating-pair formation and conjugation.
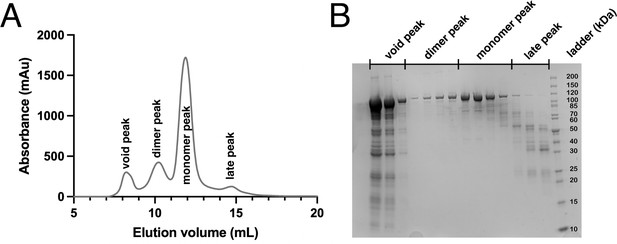
Purification of PrgB.
(A) Representative chromatogram from size-exclusion chromatography (SEC) on a Superdex 200 Increase 10/300 GL column. Four peaks are observed, corresponding to the void, PrgB dimer, PrgB monomer, and a late peak. (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) of the SEC elution fractions indicated in panel A.
-
Figure 1—figure supplement 1—source data 1
Raw image of the SDS-PAGE in Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/84427/elife-84427-fig1-figsupp1-data1-v1.zip
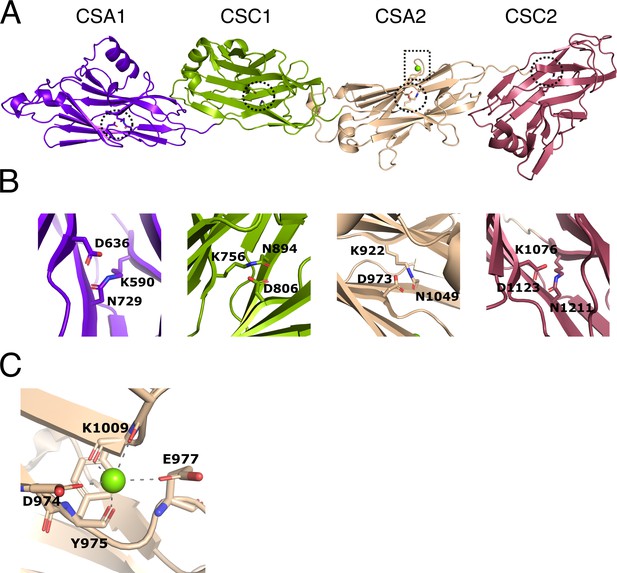
Structure of immunoglobulin (Ig)-like domains of PrgB.
(A) The Ig-like domains are arranged as tandem pairs (CSA1–CSC1 and CSA2–CSC2). (B) Each Ig-like domain has an internal isopeptide bond (indicated by the striped circle in panel A) strengthening the structural integrity of the domain. Each of the four isopeptide bonds is between a lysine and an asparagine and further stabilized by an aspartic acid residue (highlighted residues are shown in stick representation). (C) A conserved metal binding site is situated in the CSA2 domain (highlighted by a striped box in panel A), here modeled with a Mg2+ (green sphere).
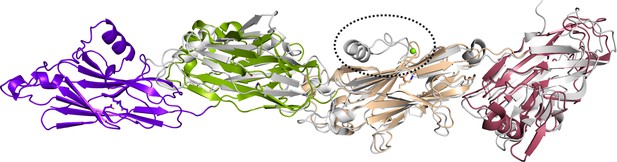
Structure of PrgB compared to the C-terminal domain of Pas (gray) (PDB code: 6E3F).
The BAR motif from Pas, missing in PrgB, is highlighted by the striped circle.
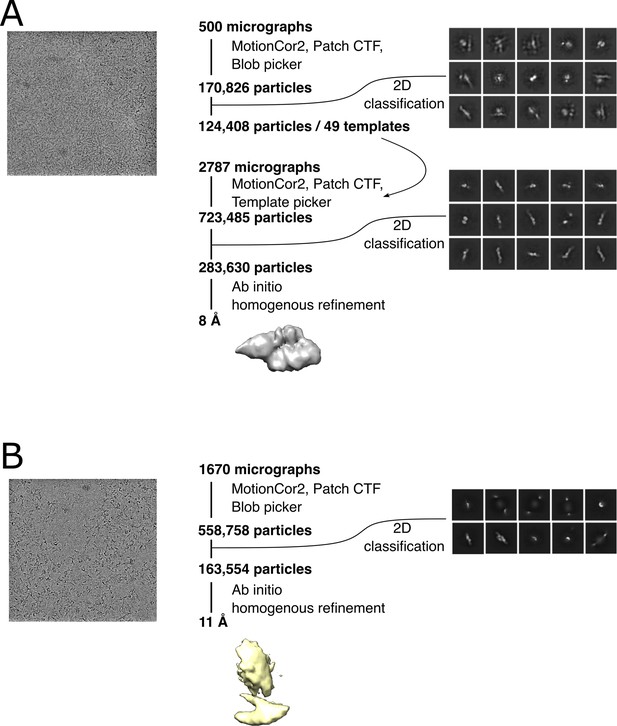
Cryo-EM data processing scheme.
Data collection and processing for the apo PrgB dataset (A) and PrgB bound to DNA (B). For both, representative micrographs and 2D classes used in the processing are shown.
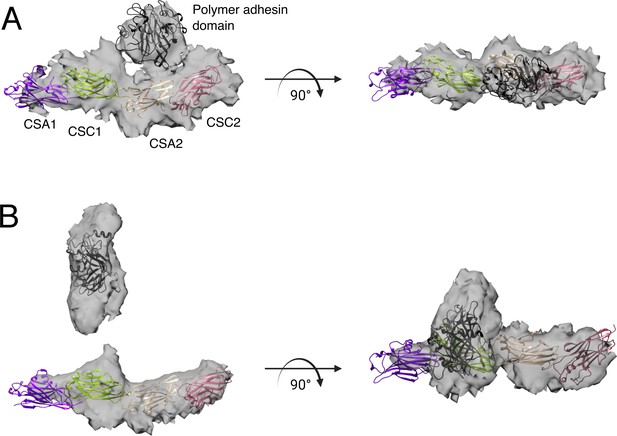
Crystal structures of the PrgB polymer adhesin domain and immunoglobulin (Ig)-like domains in cartoon representation docked into the volumes acquired by single particle cryo-EM.
(A) The PrgB188–1233 model shows the polymer adhesin domain folded back onto the Ig-like domains. (B) In the DNA-bound PrgB188–1233 model, the polymer adhesin domain has undergone a large conformation change away from the Ig-like domain.

Analysis of potential interaction of the polymer adhesin domain and stalk-like domains of PrgB.
(A) Size-exclusion chromatography of PrgB_PAD (red), PrgB_Stalk (green), and mixture (black). No shifted peaks are observed in the mixture. (B) Native polyacrylamide gel electrophoresis (PAGE) of PrgB_PAD, PrgB_Stalk, and mixture in 4–20% native gradient gel. PrgB_PAD has a strongly positively charged surface, and does not enter the gel on its own. No shift is observed when mixed with the stalk domain.
-
Figure 2—figure supplement 4—source data 1
Raw image of the SDS-PAGE in Figure 2—figure supplement 4B.
- https://cdn.elifesciences.org/articles/84427/elife-84427-fig2-figsupp4-data1-v1.zip

AlphaFold model of PrgB.
(A) Model of PrgB with N-proximal residues 157–194 in gray, the coiled-coil (COI, residues 195–260) domain in cyan, the polymer adhesin domain (PAD, residues 261–558) in black, the following linker region (residues 559–583) in orange, CSA1 (residues 584–750) in purple, CSC1 in green, CSA2 in beige, and CSC2 in red. Both N- and C-termini (residues 1–156 and 1233–1305) are removed for clarity as they were predicted to be largely unfolded. (B) The linker region between the polymer adhesin domain and the CSA1 domain, as indicated in panel A. (C) The same region as shown in panel B with the residues colored by their confidence measure (pLDDT score). Regions with a pLDDT score above 90 are expected to be modeled with high accuracy, regions with a score between 70 and 90 are expected to have a good prediction for the backbone, while regions with a pLDDT score between 50 and 70 are off low confidence. A pLDDT score <50 is a reasonable strong predictor of disorder.
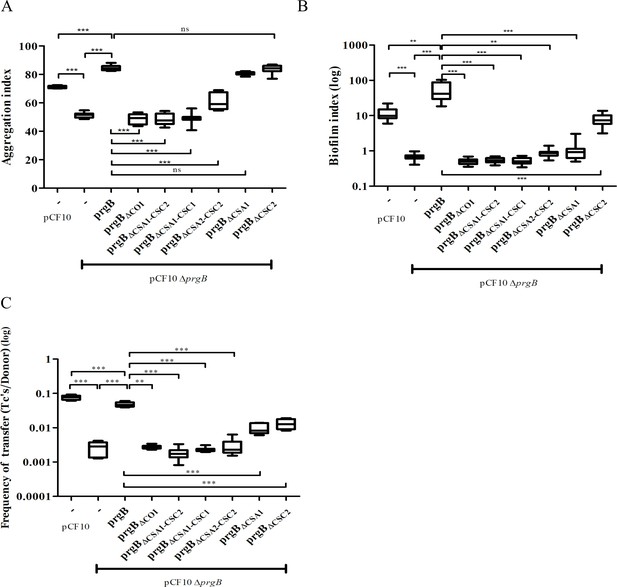
In vivo phenotypes of PrgB variants.
PrgB variants are expressed from the pMSP3545S-MCS vector in the OG1RF pCF10ΔprgB background for phenotypic analysis with three assays: (A) cellular aggregation, (B) biofilm formation, and (C) conjugation assays. For all assays, OG1RF pCF10 carrying an empty vector serves as positive control and OG1RF pCF10ΔprgB with an empty vector as negative control. The value of each column represents the average of three independent experiments and the error bars represent the standard error of the mean (SEM). Statistical significance between the PrgB variants were analyzed with one-way analysis of variance, with * indicating p < 0.05, ** indicating p < 0.01, and *** indicating p < 0.001.

Western blot showing the expression levels of the PrgB variants in E. faecalis cell wall extracts.
(A) Samples collected after 1-hr induction with cCF10 and nisin. (B) Samples collected after overnight induction with cCF10 and nisin. Estimated molecular weight of PrgB variants: WT (140 kDa); ΔCOI (137 kDa); ΔCSA1CSC2 (72 kDa); ΔCSA1CSC1 (103 kDa); ΔCSA2CSC2 (105 kDa); ΔCSA1 (121 kDa); ΔCSA2 (122 kDa).
-
Figure 3—figure supplement 1—source data 1
Raw images of SDS-PAGE.
- https://cdn.elifesciences.org/articles/84427/elife-84427-fig3-figsupp1-data1-v1.zip
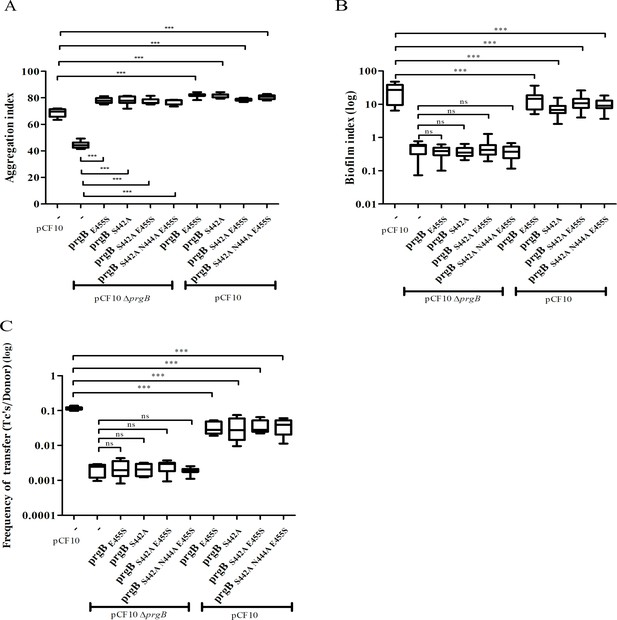
In vivo phenotypes of PrgB variants with point mutation(s) in the conserved site of the polymer adhesin domain.
PrgB variants are expressed from the pMSP3545S-MCS vector in the OG1RF pCF10ΔprgB or OG1RF pCF10 strain and analyzed in: (A) cellular aggregation, (B) biofilm formation, and (C) conjugation assays. OG1RF pCF10 carrying the empty vector serves as positive control and OG1RF pCF10ΔprgB with the empty vector as negative control. The height of each column represents the average of three independent experiments and the error bars indicate the standard error of the mean (SEM). Statistical significance between the PrgB variants were analyzed with one-way analysis of variance, with * indicating p < 0.05, ** indicating p < 0.01, and *** indicating p < 0.001.

Comparison of the DNA-binding affinity of the wild-type polymer adhesion domain from PrgB (PrgB188–1234 WT) and its S442A and N444A variant (PrgB188–1234 S442A N444A).
(A) For this mobility shift assay, protein was mixed and incubated with 50 nM DNA100 and applied to native gels. From left to right, DNA mixtures with 0, 0.16, 0.31, 0.63, 1.25, 2.5, 5, 10, or 20 mM protein were loaded and the final lane is a negative control with only 20 mM protein (and no DNA). (B) Plot of the relative intensities of the unbound (not upshifted) DNA bands normalized against the protein-free condition (lane 1). The error is the standard deviation (N = 2).
-
Figure 4—figure supplement 1—source data 1
Raw tif files of the EMSAs in Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/84427/elife-84427-fig4-figsupp1-data1-v1.zip
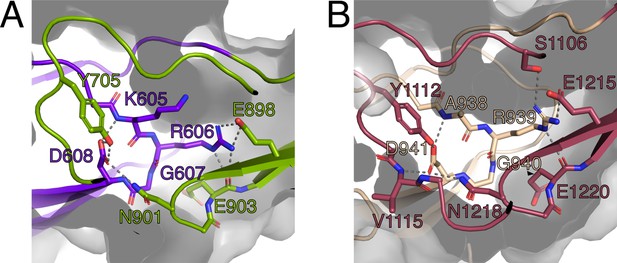
The RGD motifs play a role in the structural integrity of PrgB.
(A) Close-up view of the RGD motif in the CSA1 domain (purple). Most of the important interactions, mainly hydrogen bonds, formed by this motif are to residues on the CSC1 domain (green). (B) Close-up view of the RGD motif in the CSA2 domain (sand colored). Most of the important interactions, mainly hydrogen bonds, formed by this motif are to residues on the CSC2 domain (dark red). Potential hydrogen bonds are marked by lines and the surface of the protein indicated by transparent gray.
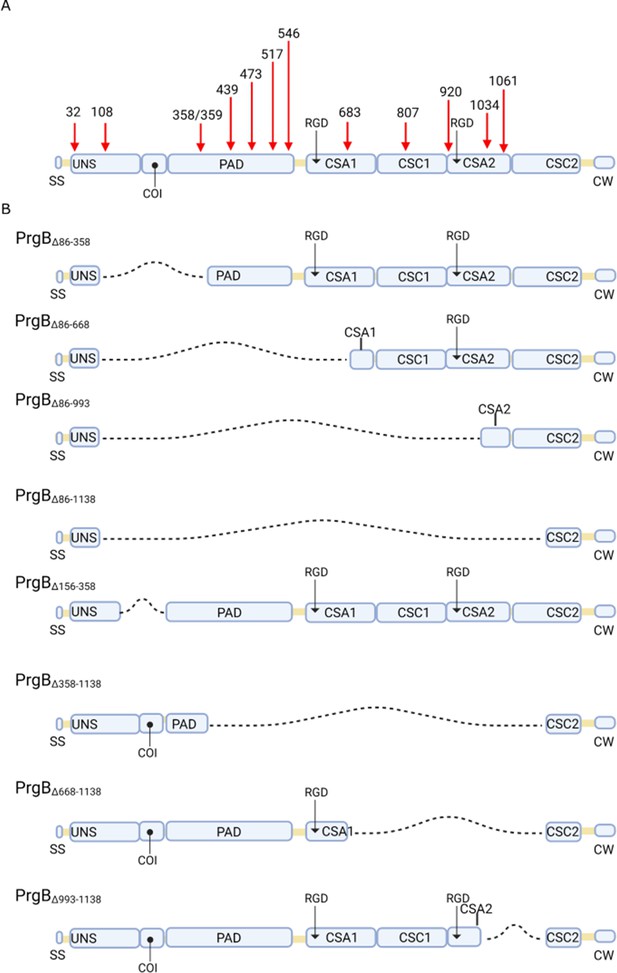
Schematic overview of previously described prgB mutants.
(A) Red arrows indicate the sites of transposon insertions that cause defective phenotypes in aggregation, biotic tissue biofilm formation, or conjugation. (B) Diagrams illustrating the specific deletion mutants discussed in the main text. Dashed lines indicate deleted regions. SS, signal sequence; UNS, unstructured region; PAD, polymer adhesin domain; COI, coiled-coil domain; CSA, cell-surface antigen; CSC, cell-surface antigen C-terminus; CW, LPXTG-containing cell wall anchor sequence.
Tables
X-ray data collection and refinement statistics.
Values within parenthesis correspond to the highest resolution shell.
| Data collection summary | PrgB584–1233 |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 76.0, 87.5, 224.8 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 47.3–1.84 (1.91–1.84) |
| Completeness (%) | 99.6 (99.5) |
| Rmeas (%) | 0.03 (0.99) |
| I/σ (I) | 12.1 (1.0) |
| CC(1/2) | 1 (0.67) |
| Redundancy | 2.0 (2.0) |
| No. unique reflections | 130,301 (12,877) |
| Refinement summary | |
| Resolution (Å) | 47.3–1.84 |
| Rwork (%) | 20.9 |
| Rfree (%) | 24.5 |
| Number of atoms | |
| Protein | 10,080 |
| Water | 816 |
| Ther ligands | 2 |
| B-factors | |
| Protein | 44.9 |
| Water | 47.7 |
| Ther ligands | 48.3 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.79 |
| Ramachandran statistics | |
| Utliers (%) | 0 |
| Allowed (%) | 1.5 |
| Favored (%) | 98.5 |
Additional files
-
Supplementary file 1
Strains, plasmids, and oligonucleotides.
- https://cdn.elifesciences.org/articles/84427/elife-84427-supp1-v1.docx
-
Supplementary file 2
Cryo-EM data collection and refinement.
- https://cdn.elifesciences.org/articles/84427/elife-84427-supp2-v1.docx
-
Supplementary file 3
Top hits of Dali search based on PDB of PrgB Ig-like domains.
- https://cdn.elifesciences.org/articles/84427/elife-84427-supp3-v1.docx
-
Supplementary file 4
Structural interpretation of previously observed PrgB phenotypes.
- https://cdn.elifesciences.org/articles/84427/elife-84427-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84427/elife-84427-mdarchecklist1-v1.docx





