Multi-centre analysis of networks and genes modulated by hypothalamic stimulation in patients with aggressive behaviours
Figures
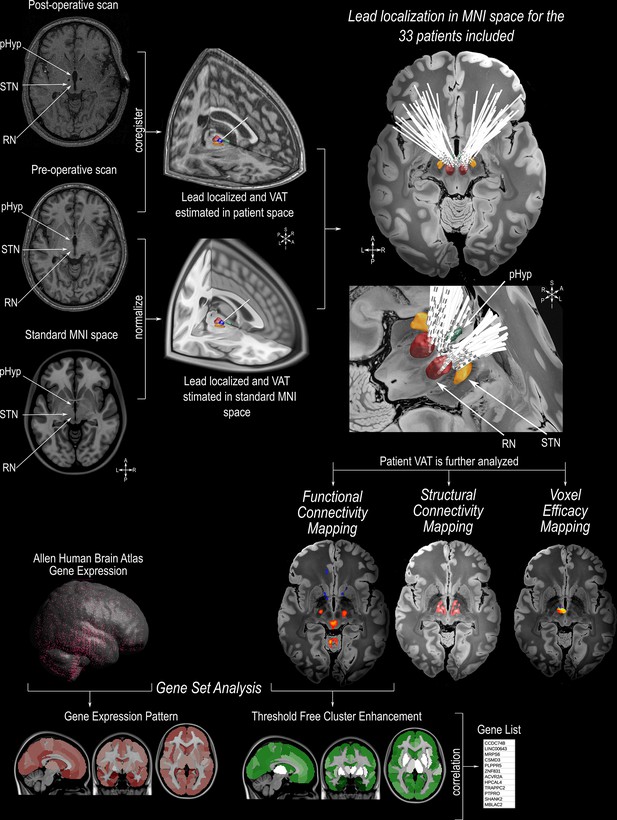
Illustration of the methodologies applied in this study.
Preoperative MRI scans were co-registered with the postoperative MRI/CT scan, followed by normalization to standard MNI152 space (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009). Individual DBS leads were manually localized in the posterior hypothalamic area (pHyp) in the patient space and normalized to MNI152 space. The estimation of the volume of activated tissue (VAT) was calculated based on individual stimulation parameters using Lead-DBS (https://www.lead-dbs.org/ See Table 1 for individual stimulation parameters). The patients’ VATs were further investigated for the analysis of the Voxel Efficacy Map (determination of the optimal stimulation site), Imaging Connectomics using Structural Connectivity Map (determining the streamlines involved in symptom improvement) and Functional Connectivity Map (determining the functionally connected areas involved in symptom improvement). For imaging Transcriptomics, we applied a Threshold Free Cluster Enhancement (TFCE) to the functional connectivity map. Functionally connected areas were averaged into the Harvard-Oxford Atlas (http://www.cma.mgh.harvard.edu/). Based on the human gene expression data from the Allen Human Brain Atlas (https://alleninstitute.org/), genes with a spatial pattern distribution similar to the TFCE map were selected for further gene ontology analysis. 3D reconstruction of the DBS leads on a 100micron resolution, 7.0 Tesla FLASH brain (https://openneuro.org/datasets/ds002179/versions/1.1.0) in MNI152 space; the pHyp label was derived from a previously published high-resolution MRI atlas of the human hypothalamic region (https://zenodo.org/record/3903588#.YHiE7pNKiF0).
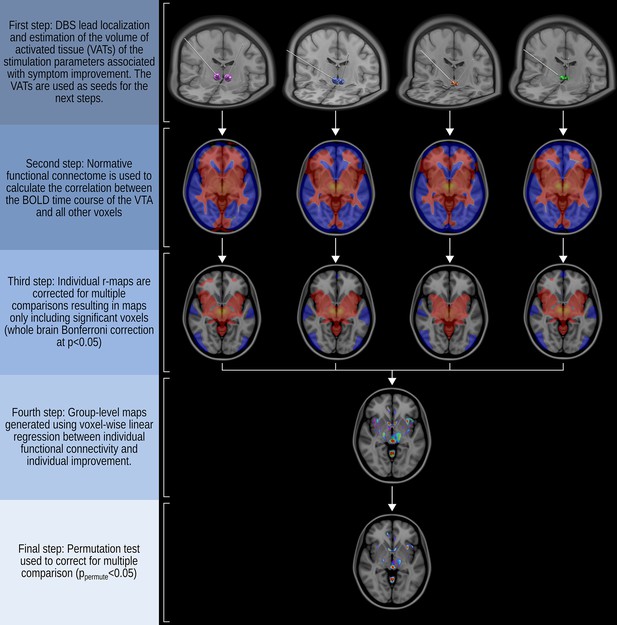
Method of generating functional connectivity maps.
This process involves the localization of the electrodes in each patient’s brain and the estimation of the volume of activated tissue (VAT) based on the stimulation parameters associated with symptom improvement. The VATs are then used as seeds for the generation of an individual r-map by correlating the BOLD time course of the VATs seed with the BOLD time course of all other voxels using the normative data of 1000subjects (Brain Genomics Superstruct Project dataset, http://neuroinformatics.harvard.edu/gsp). Individual r-maps are corrected for multiple comparisons to exclude voxels with potentially spurious correlations, resulting in an individual r-map that only included voxels surviving Bonferroni correction at the level of p<0.05. Finally, to create group-level maps, a voxel-wise linear regression analysis is performed to investigate the relationship between the functional connectivity of the VATs and the individual clinical outcome, followed by permutation correction resulting in a significant group-level functional connectivity map (ppermute<0.05). The MNI152 brain template was used for axial and three-dimensional brain images (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009).
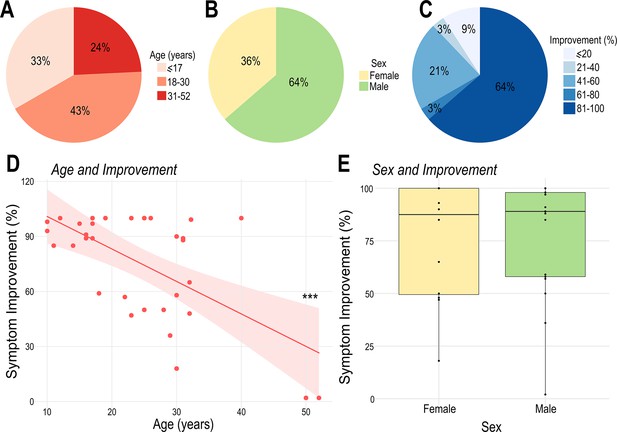
Patient demographics and treatment outcome.
(A) Patients were divided in three main groups according to age: pediatric population (≤17years, 11 out of 33), young adults (18–30years, 14 out of 33) and older adults (31–52years, 8 out of 33). (B) Distribution of males (21 out of 33) and females (12 out of 33) in this study. (C) Patient distribution according to the percentage of symptomatic improvement (≤20: 3 out of 33; 21–40: 1 out of 33; 41–60: 7 out of 33; 61–80: 1 out of 33; 81–100: 21 out of 33). Note that the majority of individuals presented over 30% improvement following treatment (criteria for being considered a treatment responder), and a large proportion of patients presented an improvement greater than 80%. (D) Age at surgery was significantly negatively correlated with postoperative symptomatic improvement (R=–0.61; R2=0.38; *** p<0.001). (E) There was no significant difference in the percentage of symptomatic improvement between male and female patients.
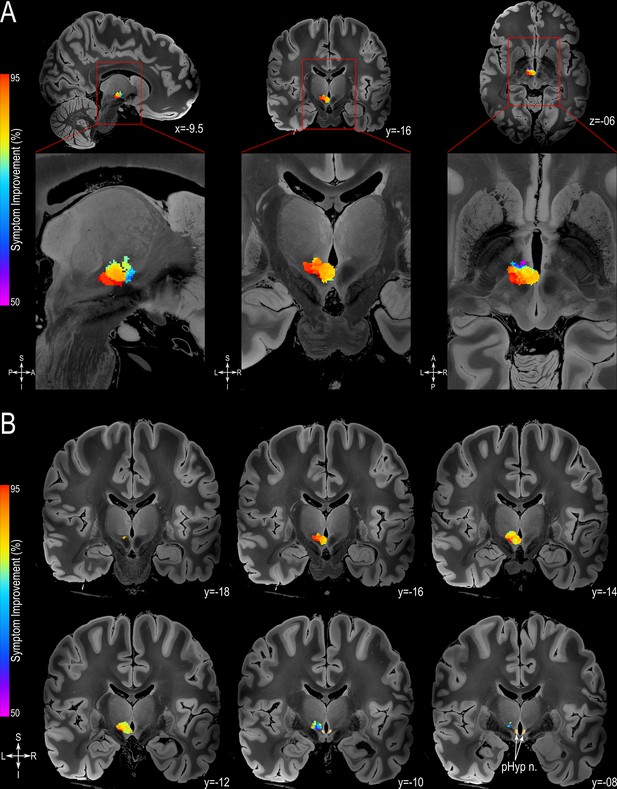
Probabilistic Sweet-spot Mapping.
(A) The area of stimulation associated with greater symptomatic improvement (red) was located in the more posterior-inferior-lateral region of the posterior hypothalamic area (from left to right: sagittal, coronal and axial views). (B) The extent of the volumes of activated tissue (VATs) responsible for eliciting at least 50% improvement is shown in successive coronal MRI slices. All results are illustrated on slices of a 100micron resolution, 7.0 Tesla FLASH brain (https://openneuro.org/datasets/ds002179/versions/1.1.0) in MNI152 space (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009). The posterior hypothalamic nucleus (pHyp n.) label (shown in beige) was derived from a previously published high-resolution MRI atlas of the human hypothalamic region (https://zenodo.org/record/3903588#.YHiE7pNKiF0).
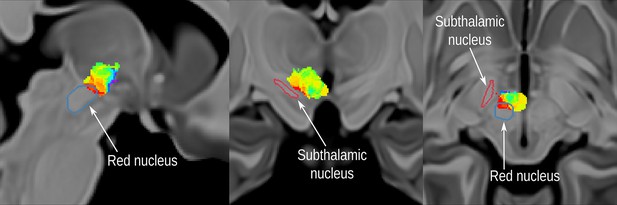
Localization of the probabilistic sweet spot mapping associated with at least 50% improvement in symptoms, in the posterior-inferior-lateral region of the posterior hypothalamic area, and in relation to the red nucleus and the subthalamic nucleus.
Note that a small portion of the map overlaps with the most superior part of the red nucleus, and no overlap with the subthalamic nucleus is observed. The labels for the Subthalamic nucleus and red nucleus are derived from a previously published high-resolution MRI atlas of the human hypothalamic region (https://zenodo.org/record/3903588#.YHiE7pNKiF0), and illustrated in the MNI152 brain (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009).
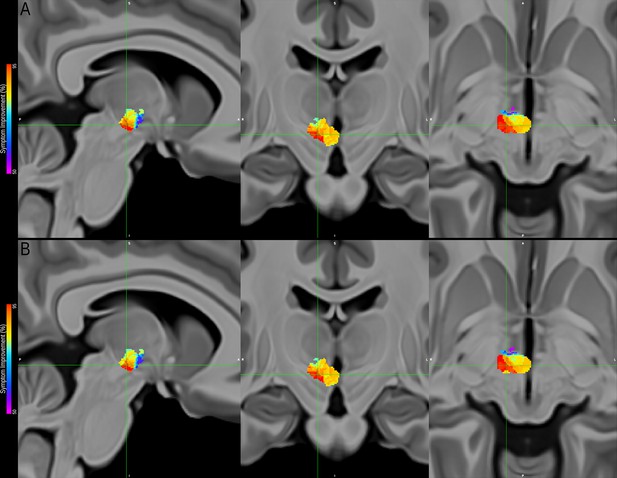
Comparison between two probabilistic sweet-spot maps performed considering amplitude (top panel, original analysis) and amplitude plus pulse width (bottom panel, additional analysis).
Note the striking similarity between maps, with the location and values of the peak corresponding to the most efficacious area for maximal symptom alleviation remaining unaltered, and only a few voxels on the periphery of the map changing in value by a couple of percentage points.
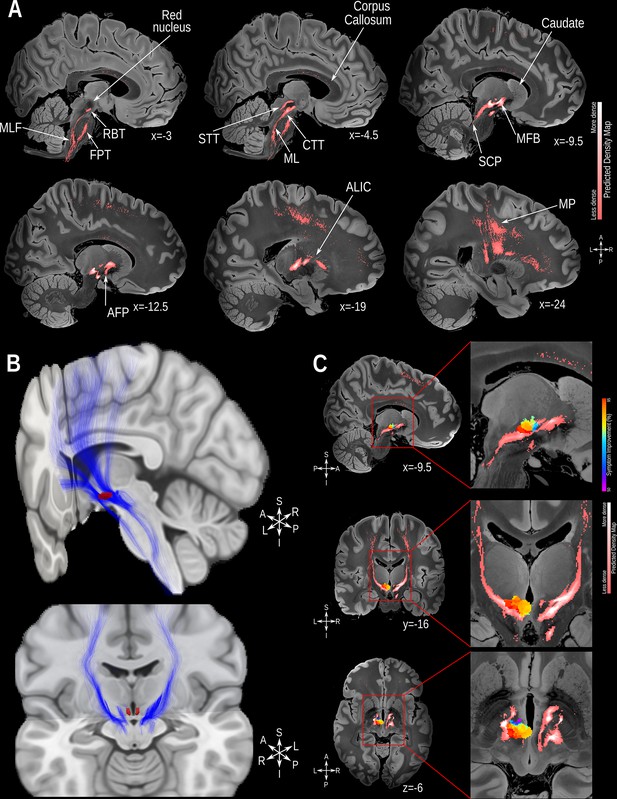
Structural connectivity mapping.
(A) Magnetic resonance imaging (MRI) in the sagittal plane showing the fiber density of streamlines connected to the volumes of activated tissue (VATs) associated with significantly greater symptomatic improvement. (B) 3D reconstruction of the streamlines associated with significantly greater improvement illustrated on the MNI152 brain (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009); the posterior hypothalamic nucleus label (in red) was derived from a previously published high-resolution MRI atlas of the human hypothalamic region (https://zenodo.org/record/3903588#.YHiE7pNKiF0). (C) MRI showing the relation between VATs responsible for eliciting at least 50% improvement and the fiber density map (from top to bottom: sagittal, coronal and axial views). The results presented in A and C are illustrated on a 100micron resolution, 7.0 Tesla FLASH brain (https://openneuro.org/datasets/ds002179/versions/1.1.0) in MNI152 space. Abbreviations: AFP: Amygdalofugal Pathway; ALIC: Anterior Limb of the Internal Capsule; CTT: Central-Tegmental Tract; FPT: Frontopontine Tract; MFB: Medial Forebrain Bundle; ML: Medial Lemniscus; MLF: Medial-Longitudinal Fasciculus; MP: Motor Projections; RBT: Rubrospinal Tract; SCP: Superior Cerebellar Peduncle; STT: Spino-Thalamic Tract.
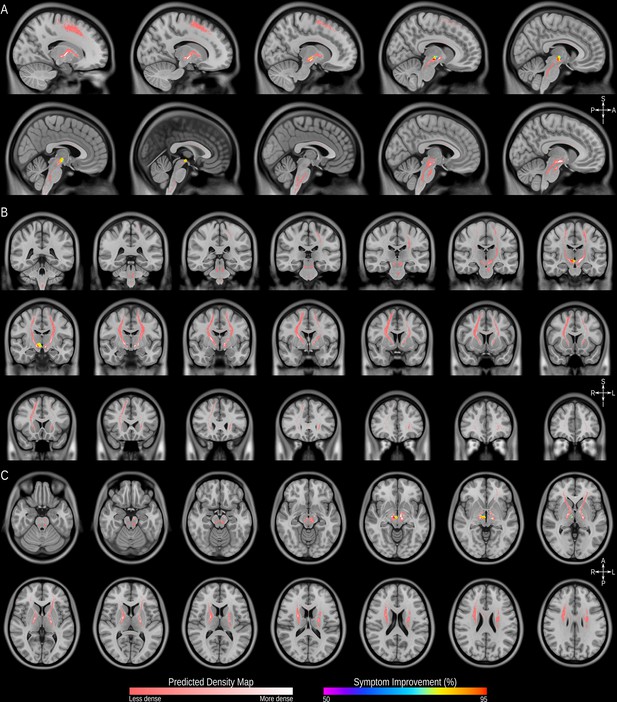
Structural connectivity mapping.
Magnetic Resonance Imaging (MRI) in the sagittal (A), coronal (B) and axial (C) planes showing the fiber density of streamlines (pink scale) connected to the volumes of activated tissue (VATs) associated with significantly greater symptom improvement and voxels associated with at least 50% improvement in symptoms (spectrum scale), illustrated in the MNI152 brain (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009).
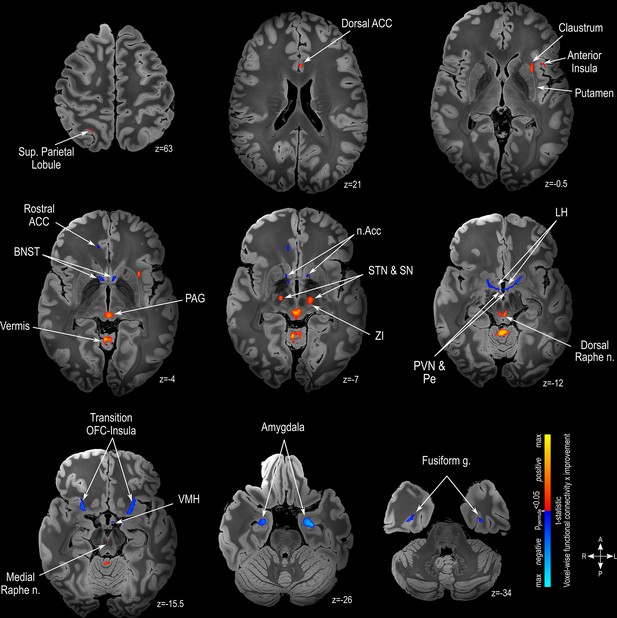
Functional connectivity mapping.
Magnetic resonance imaging (MRI) in the axial plane showing areas found to have a positive (warm colors) or a negative (cold colors) correlation between clinical outcome and functional connectivity. Results are illustrated on a 100micron resolution, 7.0 Tesla FLASH brain in MNI152 space (https://openneuro.org/datasets/ds002179/versions/1.1.0). Abbreviations: ACC: Anterior Cingulate Cortex; BNST: Bed Nucleus of Stria Teminalis; LH: Lateral Hypothalamus; n.Acc: Nucleus Accumbens; OFC: Orbitofrontal Cortex; PAG: Periaqueductal Grey matter; Pe: Periventricular Hypothalamus; PVN: Paraventricular Hypothalamus; SN: Substantia Nigra; STN: Subthalamic Nucleus; VMH: Ventromedial Hypothalamus; ZI: Zona Incerta.
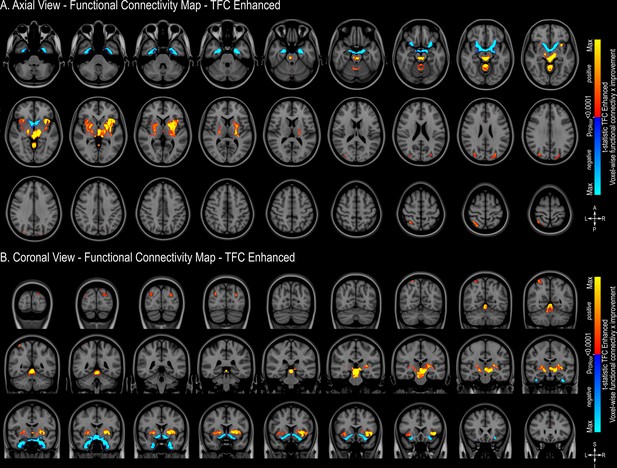
Threshold-free cluster enhancement functional connectivity mapping.
Magnetic resonance imaging (MRI) in the axial plane (A) and coronal plane (B) showing areas found to have a positive correlation between clinical outcome and functional connectivity (warm colors) or a negative correlation between outcome and functional connectivity (cold colors) FDR corrected at q<0.0001. The results are illustrated in the MNI152 brain (https://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009).
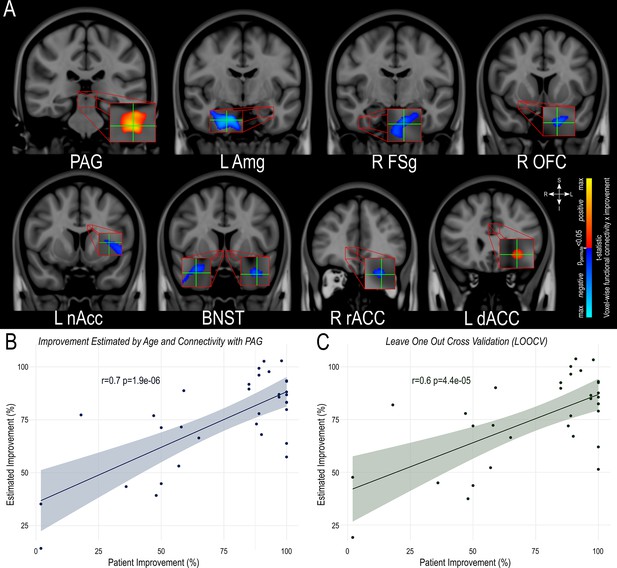
Estimation of clinical outcome.
(A) Location of the peak extracted for each area found to have significant functional connectivity with the volume of activated tissue Illustrated in the coronal plane in MNI152 standard-space (http://www.bic.mni.mcgill.ca/ServicesAtlases/HomePage). (B) A model using age and individual VAT connectivity to the periaqueductal gray significantly estimated individual symptom improvement in the whole dataset and (C) retained significance during leave-one-out cross-validation (LOOCV).
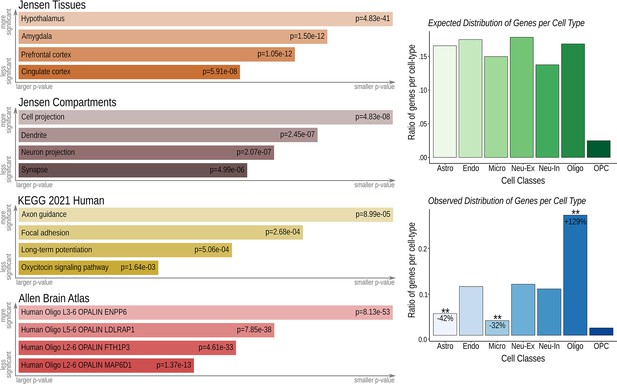
Imaging transcriptomics-gene set analysis.
The gene set analysis was performed using the TFCE-corrected distribution of clinically relevant functionally connected areas (qFDR<0.0001) along with whole brain three-dimensional expression patterns provided by the Allen Brain Atlas (http://human.brain-map.org/) Hawrylycz et al., 2012; Sunkin et al., 2013; Shen et al., 2012 averaged into the Harvard-Oxford Atlas (http://www.cma.mgh.harvard.edu/). Genes with similar spatial pattern of distribution to the functional connectivity map were Bonferroni corrected at p<0.005 and selected for further gene ontology analysis. Left panel: The EnRichr tool (https://maayanlab.cloud/Enrichr/) Kuleshov et al., 2016 was used to investigate associated biological processes, followed by specific tissue and compartment analysis provided by the Jensen Gene Ontology Tool (https://jensenlab.org/resources/proteomics/), the Kyoto Encyclopedia of Genes and Genomes (KEEG; https://www.genome.jp/kegg/) and the Allen Human Brain Atlas (http://human.brain-map.org/). Right panel: A cell-specific aggregate gene set provided by Seidlitz et al., 2020 was used to determine the cell types associated with these genes. Results were confirmed to be non-random using permutation testing (1000 permutations, ** p<0.01).
Tables
Demographics.
| Case | Sex | Age range | Improvement (%) | Laterality | DBS System | Stimulation Settings | |
|---|---|---|---|---|---|---|---|
| Right | Left | ||||||
| 1 | M | 31–52 | 100 | Bilateral | Medtronic 3387 | 1.8V; 180Hz; 60msec | 1.8V; 180Hz; 60msec |
| 2 | M | ≤17 | 97 | Bilateral | Medtronic 3387 | 2.2V; 200Hz; 90msec | 2.2V; 200Hz; 90msec |
| 3 | M | ≤17 | 98 | bilateral | Medtronic 3387 | 2.5V; 180Hz; 90msec | 2.5V; 180Hz; 90msec |
| 4 | M | ≤17 | 89 | Bilateral | Medtronic 3387 | 5.0V; 210Hz; 90msec | 5.0V; 210Hz; 90msec |
| 5 | F | ≤17 | 93 | Bilateral | Medtronic 3387 | 2.0V; 200Hz; 90msec | 2.0V; 200Hz; 90msec |
| 6 | F | ≤17 | 85 | Bilateral | Medtronic 3387 | 4.0V; 180Hz; 90msec | 4.0V; 180Hz; 90msec |
| 7 | F | ≤17 | 100 | Bilateral | Medtronic 3387 | 3.5V; 180Hz; 90msec | 3.5V; 180Hz; 90msec |
| 8 | F | 31–52 | 100 | Bilateral | Medtronic 3387 | 5.0V; 200Hz; 100msec | 5.0V; 200Hz; 100msec |
| 9 | F | 18–30 | 100 | Bilateral | Medtronic 3387 | 2.3V; 200Hz; 120msec | 2.3V; 200Hz; 120msec |
| 10 | M | ≤17 | 85 | Bilateral | Medtronic 3387 | 2.0V; 130Hz; 60msec | 2.0V; 130Hz; 130msec |
| 11 | M | 18–30 | 100 | Bilateral | Medtronic 3387 | 3.0V; 180Hz; 90msec | 3.0V; 180Hz; 90msec |
| 12 | M | ≤17 | 89 | Bilateral | Medtronic 3387 | 5.5V; 185Hz; 130msec | 5.5V; 185Hz; 130msec |
| 13 | F | 18–30 | 47 | Bilateral | Medtronic 3387 | 7.0V; 250Hz; 120msec | 7.0V; 250Hz; 120msec |
| 14 | M | 18–30 | 100 | Bilateral | Medtronic 3387 | 5.0V; 210Hz; 130msec | 5.0V; 210Hz; 130msec |
| 15 | F | 18–30 | 90 | Bilateral | Medtronic 3387 | 3.5V; 180Hz; 90msec | 3.5V; 180Hz; 90msec |
| 16 | M | 18–30 | 100 | Bilateral | Medtronic 3387 | 5.0V; 200Hz; 100msec | 5.0V; 200Hz; 100msec |
| 17 | F | ≤17 | 100 | Bilateral | Medtronic 3387 | 3.5V; 180Hz; 90msec | 3.5V; 180Hz; 90msec |
| 18 | M | ≤17 | 97 | Bilateral | Medtronic 3387 | 4.5V; 180Hz; 90msec | 4.5V; 180Hz; 90msec |
| 19 | M | 31–52 | 88 | Bilateral | Medtronic 3389 | 3.0V; 180Hz; 90msec | 3.0V; 180Hz; 90msec |
| 20 | M | ≤17 | 91 | Bilateral | Medtronic 3389 | 3.2V; 180Hz; 90msec | 3.2V; 180Hz; 90msec |
| 21 | F | 18–30 | 18 | Bilateral | Medtronic 3389 | 3.8V; 180Hz; 90msec | 3.8V; 180Hz; 90msec |
| 22 | M | 31–52 | 89 | Bilateral | Medtronic 3389 | 3.5V; 180Hz; 90msec | 3.5V; 180Hz; 90msec |
| 23 | M | 31–52 | 2 | Bilateral | Medtronic 3389 | 4.5V; 150Hz; 247msec | 4.5V; 150Hz; 247msec |
| 24 | M | 18–30 | 57 | Unilateral | Medtronic 3389 | 0.3V; 150Hz; 450msec | Not applicable |
| 25 | M | 31–52 | 2 | Unilateral | Medtronic 3389 | 0.9V; 150Hz; 450msec | Not applicable |
| 26 | F | 31–52 | 65 | Bilateral | Medtronic 3389 | 0.1V; 60Hz; 180msec | 0.1V; 60Hz; 300msec |
| 27 | M | 18–30 | 59 | Bilateral | Medtronic 3389 | 0.7V; 150Hz; 330msec | 0.5V; 150Hz; 450msec |
| 28 | F | 31–52 | 48 | Unilateral | Medtronic 3389 | 0.1V; 150Hz; 450msec | Not applicable |
| 29 | M | 18–30 | 100 | Bilateral | Medtronic 3387 | 2.0V; 180Hz; 120msec | 2.0V; 180Hz; 120msec |
| 30 | M | 18–30 | 50 | Bilateral | Boston, Vercise | 1.0mA; 185Hz; 90msec | 1.0mA; 185Hz; 90msec |
| 31 | F | 18–30 | 50 | Bilateral | Boston, Vercise | 1.2mA; 113Hz; 120msec | 1.2mA; 113Hz; 120msec |
| 32 | M | 18–30 | 36 | Bilateral | Boston, Vercise | 1.0mA; 170Hz; 70msec | 1.0mA; 170Hz; 70msec |
| 33 | M | 18–30 | 58 | Bilateral | Boston, Vercise | 3mA; 185Hz; 60msec | 3mA; 185Hz; 60msec |
| To preserve patients' anonymization, the diagnoses observed in this group are presented as the following list, from more to less frequent. Epilepsy, autism spectrum disorder, tuberous sclerosis, congenital rubella, intermittent explosive disorder, agenesia of the corpus callosum, schizophrenia, obsessive-compulsive disorder, West syndrome, Landau-Kleffner syndrome, Cri-du-chat syndrome, Lennox-Gastaut syndrome, Sotos syndrome, meningoencephalitis, perinatal hypoxia, periventricular leucomalacia, microcephaly, arteriovenous malformation. | |||||||
Estimation of clinical outcome based on functional connectivity map and patient age.
| Functionally connected brain area | Peak coordinate | R | R2 | p-value |
|---|---|---|---|---|
| Periaqueductal Grey Matter | x=-1 y=-30 z=-10 | 0.725 | 0.525 | 1.86e-06 |
| Vermis | x=1 y=-49 z=-12 | 0.702 | 0.493 | 5.21e-06 |
| Medial Raphe nucleus | x=0 y=-25 z=-15 | 0.689 | 0.475 | 9.11e-06 |
| Right Subst. Nigra, Subthalamic n., Zona Incerta | x=16 y=-14 z=-7 | 0.681 | 0.464 | 1.28e-05 |
| Left Subst. Nigra, Subthalamic n., Zona Incerta | x=-14 y=-16 z=-7 | 0.673 | 0.453 | 1.77e-05 |
| Left Claustrum | x=-30 y=14 z=-2 | 0.672 | 0.451 | 1.88e-05 |
| Left Amygdala | x=-25 y=-8 z=-27 | 0.672 | 0.451 | 1.88e-05 |
| Right Fusiform Gyrus | x=37 y=-10 z=-34 | 0.668 | 0.447 | 2.14e-05 |
| Left Putamen | x=-33 y=-4 z=2 | 0.656 | 0.430 | 3.41e-05 |
| Left Dorsal Anterior Cingulate Cortex | x=-2 y=25 z=22 | 0.654 | 0.428 | 3.65e-05 |
| Right Superior Parietal Lobule | x=23 y=-62 z=63 | 0.652 | 0.425 | 3.95e-05 |
| Left Transition Orbitofrontal Cortex- Insula | x=-24 y=11 z=-18 | 0.645 | 0.416 | 5.06e-05 |
| Right Nucleus Accumbens | x=11 y=8 z=-7 | 0.645 | 0.416 | 5.07e-05 |
| Right Amygdala | x=-22 y=-6 z=-26 | 0.645 | 0.416 | 5.14e-05 |
| Left Anterior Insula | x=-39 y=18 z=-1 | 0.642 | 0.413 | 5.59e-05 |
| Right Rostral Anterior Cingulate Cortex | x=9 y=40 z=-5 | 0.638 | 0.407 | 6.57e-05 |
| Right Bed Nucleus Of The Stria Terminallis | x=7 y=8 z=-5 | 0.638 | 0.407 | 6.57e-05 |
| Left Bed Nucleus Of The Stria Terminallis | x=-6 y=8 z=-5 | 0.638 | 0.407 | 6.57e-05 |
| Left Nucleus Acuumbens | x=-11 y=8 z=-7 | 0.638 | 0.407 | 6.57e-05 |
| Right Transition Orbitofrontal Cortex- Insula | x=25 y=10 z=-15 | 0.638 | 0.407 | 6.57e-05 |
| Right Hypothalamus | x=5 y=-3 z=-11 | 0.631 | 0.398 | 8.25e-05 |
| Left Fusiform Gyrus | x=-34 y=-12 z=-33 | 0.629 | 0.396 | 8.72e-05 |
| Left Hypothalamus | x=-4 y=-3 z=-11 | 0.621 | 0.386 | 1.13e-04 |
Additional files
-
Supplementary file 1
Matrix of the correlations between estimated symptom improvement (i.e. linear model of age and functional connectivity of the two areas) and the measured improvement.
- https://cdn.elifesciences.org/articles/84566/elife-84566-supp1-v2.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84566/elife-84566-mdarchecklist1-v2.pdf





