The mutational signatures of poor treatment outcomes on the drug-susceptible Mycobacterium tuberculosis genome
Figures
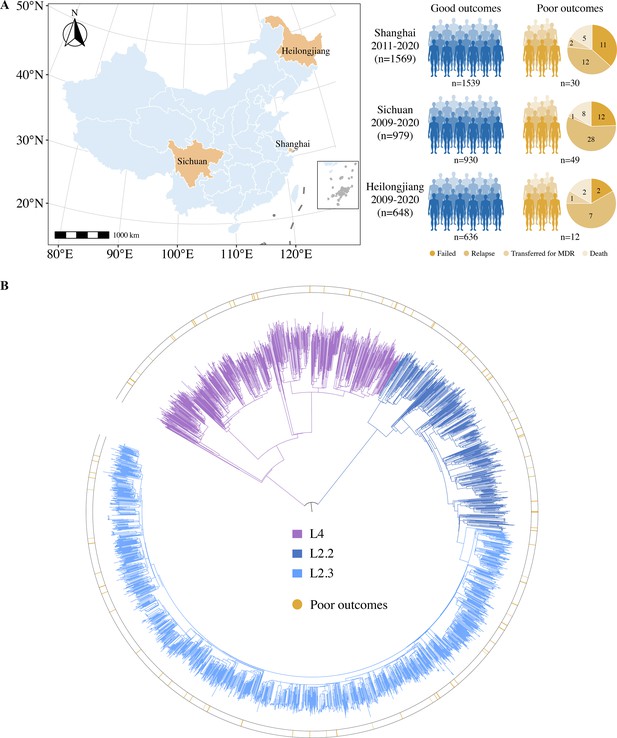
Sample origin and genetic structure of Mycobacterium tuberculosis.
(A) Geographic location of the samples analyzed and study cohort characteristics. (B) The phylogenetic tree of 3196 drug-susceptible tuberculosis strains. The different colors on the branches indicate different lineages and sublineages. The outside circle indicates the treatment outcomes of corresponding patients.
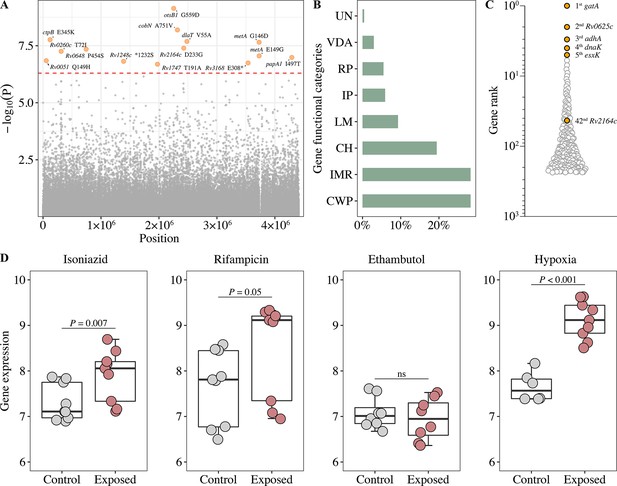
Generation of the functional mutation set.
(A) Manhattan plots of genome-wide association study (GWAS) for fixed single nucleotide polymorphisms (SNPs) associated with poor treatment outcomes. The dashed red line highlights the Bonferroni-corrected threshold (p=5.04 × 10–7). (B) Distribution of GWAS identified unfixed SNPs across gene functional categories. CWP, cell wall, and cell processes; IMR, intermediary metabolism, and respiration; CH, conserved hypotheticals; LM, lipid metabolism; IP, information pathways; RP, regulatory proteins; VDA, virulence, detoxification, adaptation; UN, unknown. (C) Gene prioritization strategies (based on p-value rank) for significantly associated unfixed SNPs. (D) Gene expression from RNA-seq (log2FPKM) of Rv2164c under drug pressure and hypoxia.
-
Figure 2—source data 1
GWAS identified fixed SNPs.
- https://cdn.elifesciences.org/articles/84815/elife-84815-fig2-data1-v2.docx
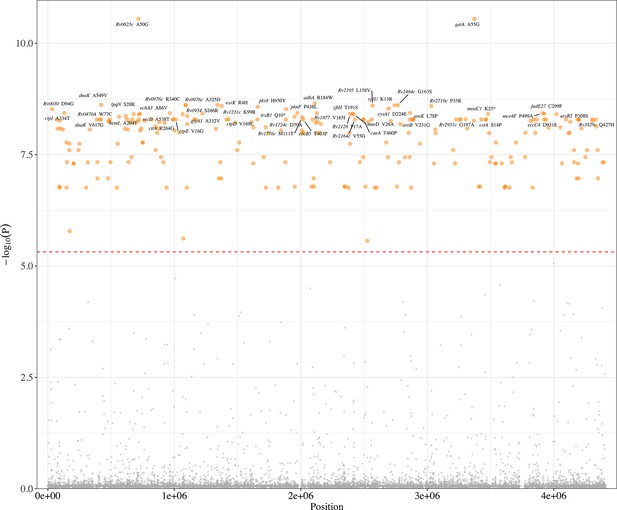
Manhattan plots of unfixed single nucleotide polymorphisms (SNPs) associated with poor treatment outcomes.
The top 50 unfixed mutations were annotated with the gene. The dashed red line highlights the Bonferroni-corrected threshold (p=4.82 × 10–6).
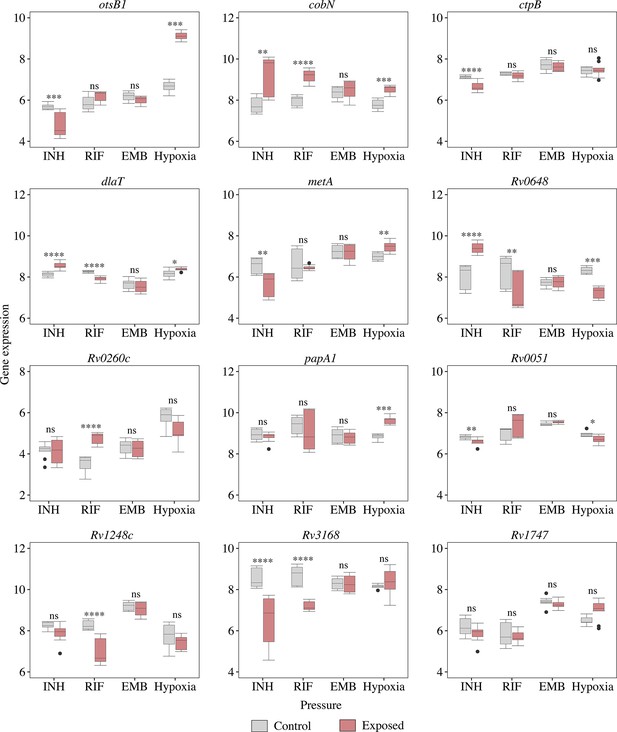
Gene expression (log2FPKM) from RNA-seq after drug exposure and hypoxia.
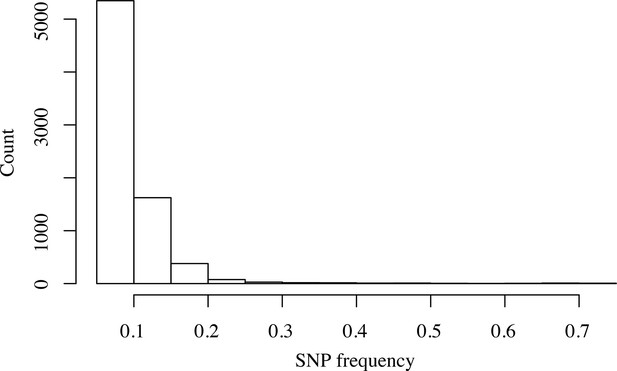
Within-host frequency distribution of genome-wide association study (GWAS)-identified unfixed mutations.
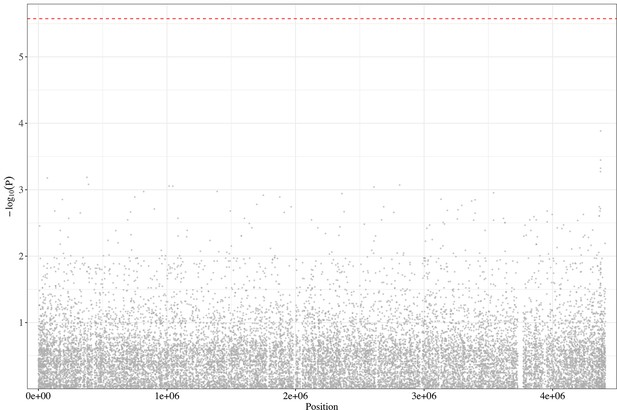
Manhattan plot of genome-wide association study (GWAS) analysis based on the Malawi dataset.
The dashed red line highlights the Bonferroni-corrected threshold (p=2.66 × 10–6).
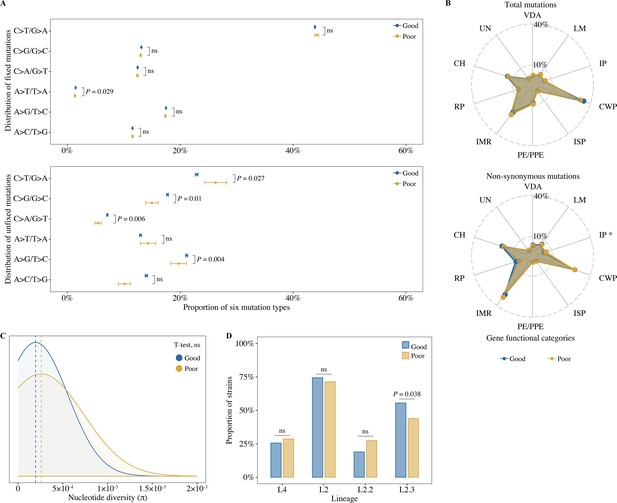
Bacterial whole-genome mutation features between patients with different treatment outcomes.
(A) The proportion of six mutation types in all fixed and unfixed mutations (t-test, mean range: mean ± SE). (B) Distribution of total unfixed mutations and nonsynonymous unfixed mutations across gene functional categories (t-test). VDA, virulence, detoxification, adaptation; LM, lipid metabolism; IP, information pathways; CWP, cell wall, and cell processes; ISP, insertion seqs and phages; IMR, intermediary metabolism, and respiration; RP, regulatory proteins; CH, conserved hypotheticals; UN, unknown. (C) Comparison of nucleotide genetic diversity between isolated patients with good and poor outcomes (t-test). (D) Distribution of Mycobacterium tuberculosi (MTB) lineages and sublineages (chi-square test). p-value <0.05 was considered significant. *, p<0.05, ns, no significant.
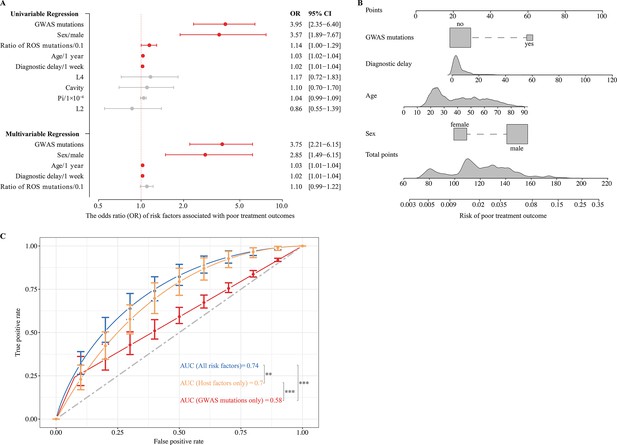
Effects of genome-wide association study (GWAS) identified mutations on tuberculosis treatment outcomes.
(A) Univariable and multivariable logistic regression on the risk factors for poor treatment outcomes. (B) Nomogram for predicting the probability of poor treatment outcomes. (C) ROC curves are based on risk factors that may be predictive of tuberculosis treatment outcomes. p-value <0.05 was considered significant. *p<0.05, **p<0.01, ***p<0.001.
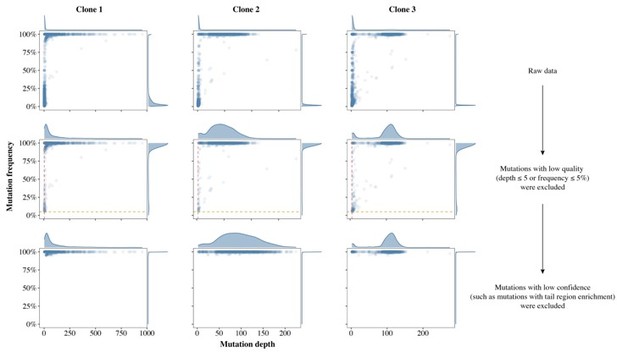
Schematics of false positive filter in three single colonies.
The density plot above the scatter plot shows the distribution of mutation depth while the plot to the right of the scatter plot shows the distribution of mutation frequency. The first row shows the SNP calling results from the raw data, in which there were many false positive mutations (FPMs). The second row shows the results after most FPMs were filtered out, leaving only those SNPs with frequency greater than 5% (horizontal yellow dashed line) and sequencing depth greater than 5 (vertical red dashed line). The third row shows the results after the remaining FPMs were filtered out with our validated pipeline.
Tables
Comparison of the ratio of strains carrying the GWAS-identified fixed mutation in different lineages.
| L2 | L4 | P-value | L2 | L4 | P-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rv0051 Q149H | Yes | 5 | 5 | 0.13F | ctpB E345K | Yes | 27 | 15 | 0.14 | |
| No | 2368 | 818 | No | 2346 | 808 | |||||
| Rv0260c T72I | Yes | 41 | 22 | 0.09 | Rv0648 P454S | Yes | 87 | 42 | 0.07 | |
| No | 2332 | 801 | No | 2286 | 781 | |||||
| Rv1248c *1232S | Yes | 87 | 43 | 0.05 | Rv1747 T191A | Yes | 94 | 66 | <0.001 | |
| No | 2286 | 780 | No | 2279 | 757 | |||||
| otsB1 G559D | Yes | 14 | 9 | 0.14 | cobN A751V | Yes | 98 | 60 | <0.001 | |
| No | 2359 | 814 | No | 2275 | 763 | |||||
| Rv2164c D233G | Yes | 96 | 46 | 0.06 | dlaT V55A | Yes | 94 | 50 | 0.01 | |
| No | 2277 | 777 | No | 2279 | 773 | |||||
| Rv3168 E308* | Yes | 17 | 9 | 0.30 | metA G146D | Yes | 106 | 50 | 0.07 | |
| No | 2356 | 814 | No | 2267 | 773 | |||||
| metA E149G | Yes | 104 | 56 | 0.01 | papA1 I497T | Yes | 5 | 1 | 1F | |
| No | 2273 | 763 | No | 2368 | 822 |
Comparison of the ratios of strains carrying at least one GWAS-identified fixed mutation from relapse cases with strains from all other patients.
| GWAS-identified mutations | P-value | |||
|---|---|---|---|---|
| Yes | No | |||
| Relapse | Yes | 13 | 34 | P < 0.001 |
| No | 241 | 2908 |
Comparison of the ratio of strains carrying at least one GWAS-identified fixed mutation from the relapse cases with strains from patients with other poor treatment outcomes.
| GWAS-identified mutations | P-value | |||
|---|---|---|---|---|
| Yes | No | |||
| Relapse | Yes | 13 | 34 | P = 0.422 |
| No | 9 | 35 |





