High-throughput library transgenesis in Caenorhabditis elegans via Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS)
Figures
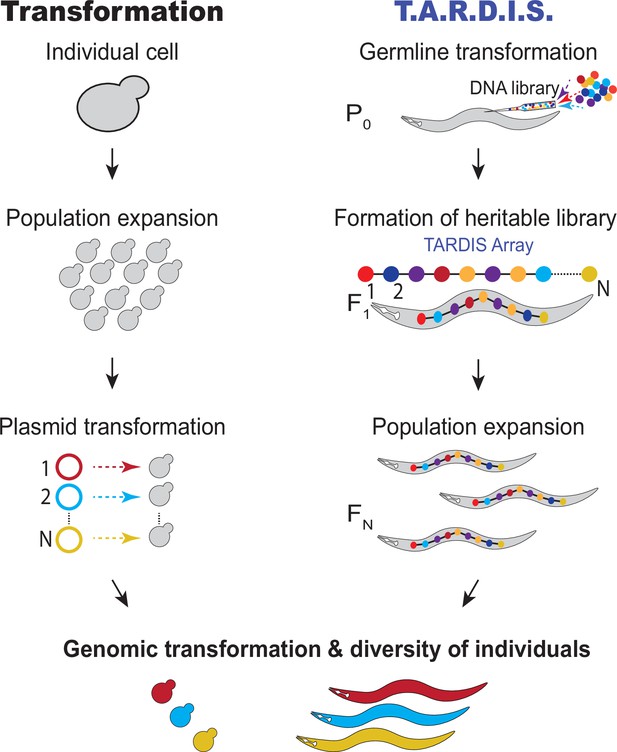
Transformation compared to Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS).
For transformation, a large population of cells are individually transformed with a DNA library, resulting in a diverse population of individuals. TARDIS achieves a diversity of individuals by splitting transgenesis into two separate processes: (1) the introduction of a diverse library, which is formed into a TARDIS library array, passed down to future generations and thus replicated; and (2) an event that triggers the integration a sequence from the library at random, resulting in a diversity of integrated sequences.
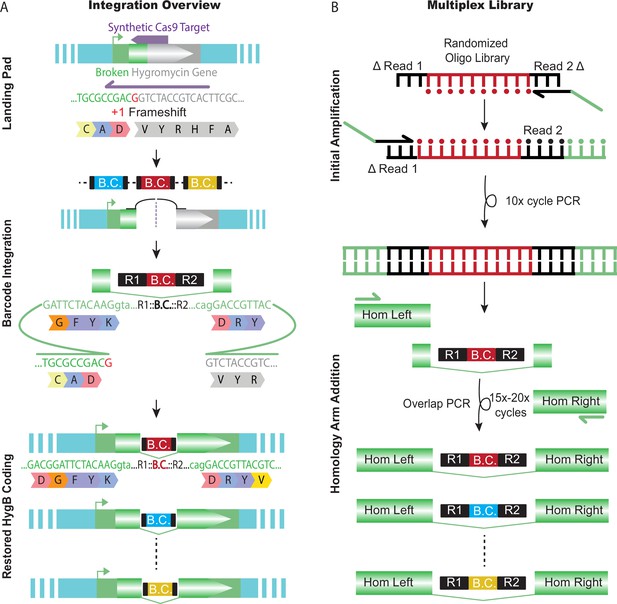
Barcode landing pad and diverse donor library.
(A) Schematic design for the barcode landing pad and integration. A broken hygromycin resistance gene is targeted by Cas9, which repairs off the Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) array, integrating a barcode and restoring the functionality of the gene. (B) The TARDIS multiplex library was created from a randomized oligo library, which underwent 10 cycles of PCR to make a dsDNA template. The barcode fragment was then added into a three fragment overlap PCR to add homology arms and make the final library for injection.
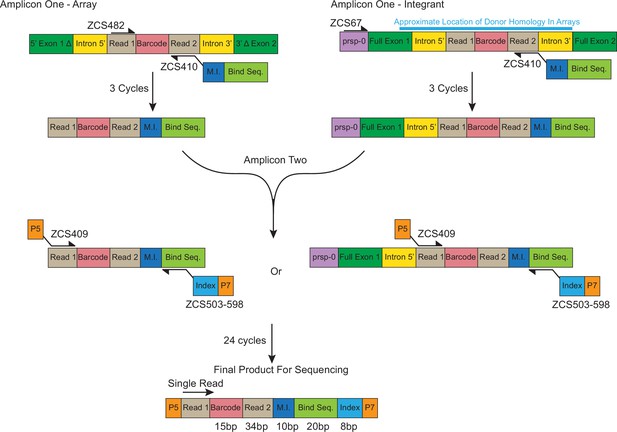
Schematic layout for the two separate PCR processes for identification of barcode counts in arrays (Amplicon One-Array) and integrants (Amplicon One-Integrant).
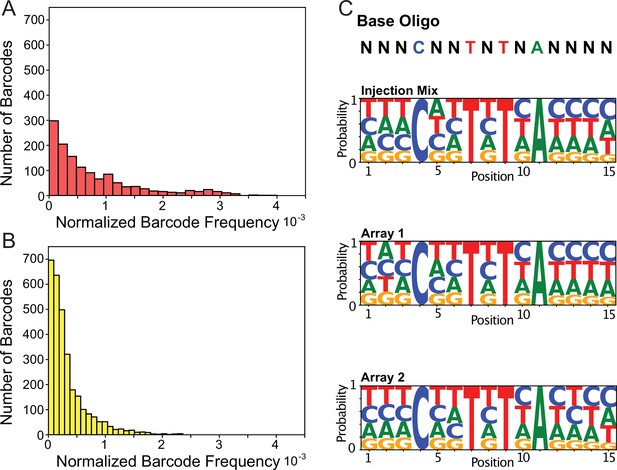
Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) library arrays can contain large barcode diversity.
(A) Frequency distribution of 1319 unique barcodes in array 1 (PX816). (B) Frequency distribution of the 3001 unique barcode sequences in array 2 (PX817). (C) Sequence logo probabilities of the 15 base pair positions of the barcodes in the injection mix, array 1 and array 2.
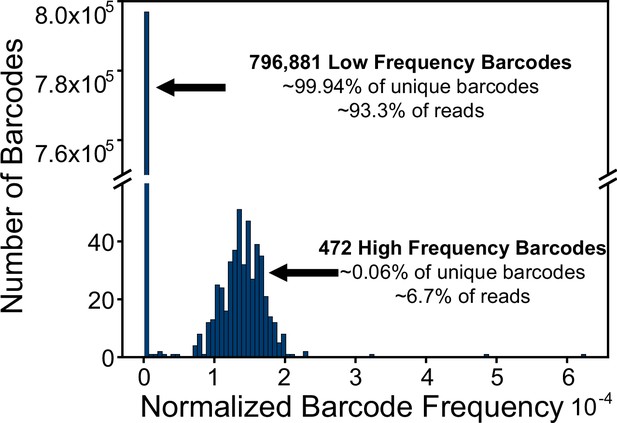
Barcode frequency in injection mix.
Barcode frequencies for Injection mix used for arrays 1–3. There are approximately 1 million reads represented. In total, 797,353 unique sequences were identified. A few of these unique sequences were represented at a higher frequency with a count cutoff greater than 50.
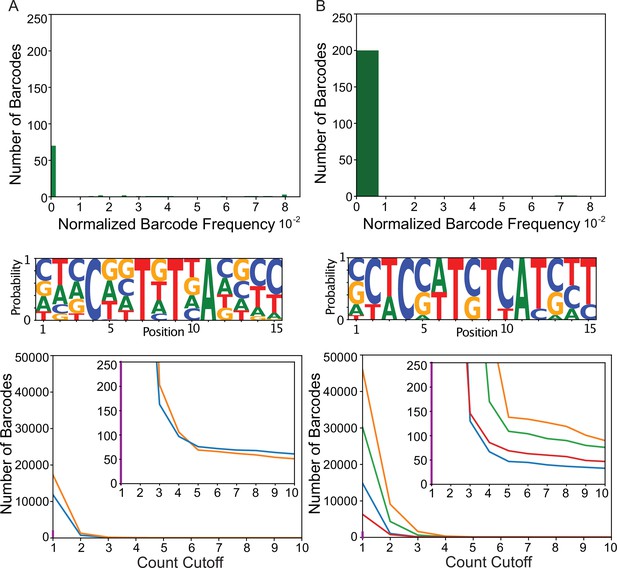
Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) array 3.
Two individual arrays were isolated from the same plate. Both show considerably less diversity than TARDIS arrays 1 and 2. Distribution of unique barcode frequencies, sequence logo base pair probability, and count cutoff for (A) TARDIS array 3 profile 1 and (B) TARDIS array 3 profile 2.
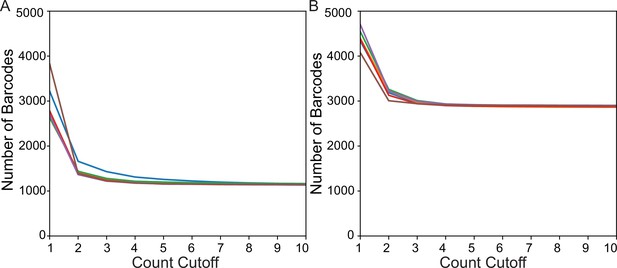
Determination of proper count cutoff for (A) Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) array 1 and (B) TARDIS array 2.
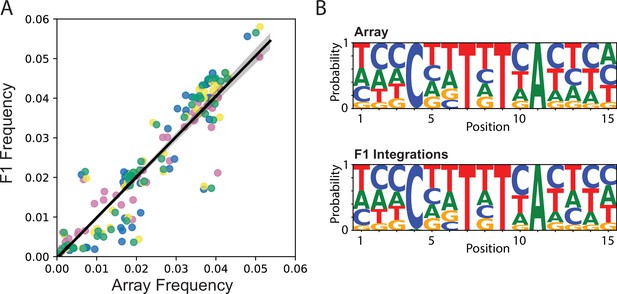
Integration frequency from Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) library array to F1.
(A) Frequency of integration from TARDIS library array to the F1, R ≈ 0.96, p≈5.7 × 10-154. Different colors represent four biological replicates. Line shading represents 95% confidence interval. (B) Sequence probabilities of PX786 compared to the F1 integrations (91 unique barcodes were identified in the array and 118 in the F1s, with a five read threshold).
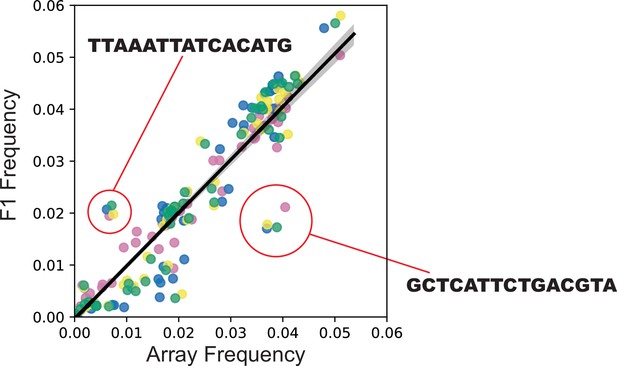
F1 integration events followed a consistent pattern, with replicated outlier barcode sequences.
Generally, the same barcodes integrated at approximately the same frequency across the four replicates.
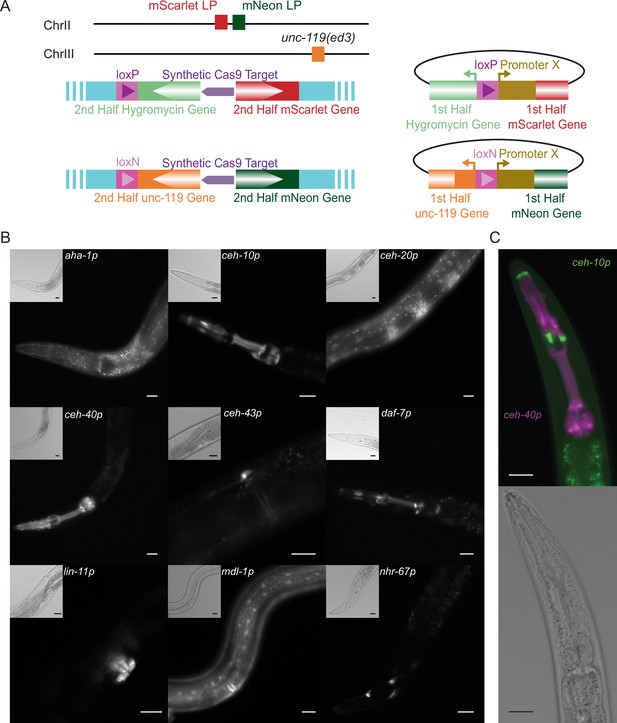
Transgenic Arrays Resulting in Diversity of Integrated Sequences (TARDIS) promoter library.
(A) Overview of the two split landing pads and their associated promoter insertion vectors. Both the selective marker and the fluorophore expression are restored upon correct integration. (B) Transcriptional reporters for nine genes were recovered from a single heatshock of a single TARDIS array line (PX819). Integration was into the single mScarlet-I/HygR landing pad. Main images show mScarlet-I expression for the indicated reporter while insets show polarized image of the same region. (C) Example simultaneous, dual integration from a single TARDIS array into the double landing pad strain with PEST. ceh-10p::mNeonGreen::PEST is false-colored green and ceh-40p::mScarlet-I::PEST is false-colored magenta. All scale bars represent 20 µm.
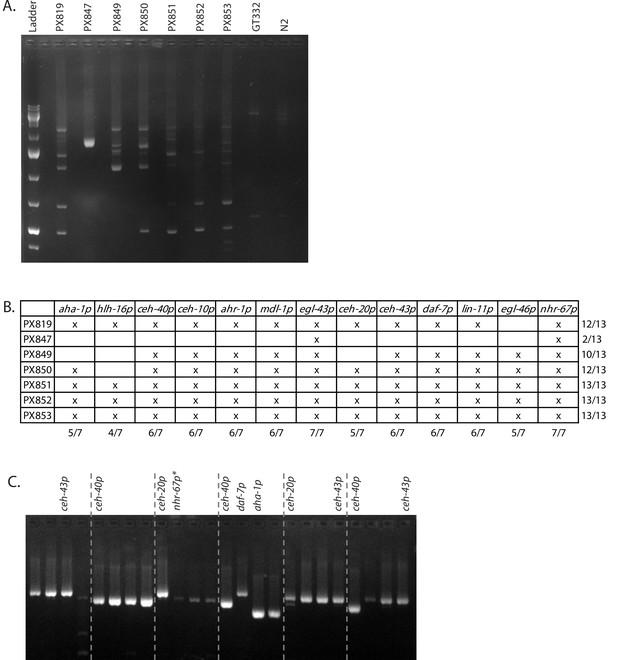
Transformation efficiency for promoter arrays.
(A) Promoters from seven array-bearing lines were amplified using universal primers and show distinct profiles. Note that not all promoters found in the arrays were detected in this screen. (B) Promoters contained in each array-bearing line as determined by promoter specific PCR (C) Line PX819 was heat-shocked to trigger integration and four hygromycin-resistant progeny were singled from each of the 59/60 plates with hygromycin-resistant individuals. Singled worms were screened by PCR, and select promoters were chosen for sequencing based on their size profile. PCR and sequencing result are shown for representative plates. *Due to their size of the egl-46 and nhr-67, promoters do not reliably amplify with the universal primers. Therefore, samples with no or weak amplification were rescreened with primers specific to these two promoters (not shown).
-
Figure 5—figure supplement 1—source data 1
Raw image files for results shown in Figure 5.
- https://cdn.elifesciences.org/articles/84831/elife-84831-fig5-figsupp1-data1-v1.zip
Tables
Characteristics of injected promoters and presence in tested array line (PX819) and integrated lines derived from that array.
| Promoter | Promoter size (bp) | Expected expression location | Array | Integrated |
|---|---|---|---|---|
| aha-1 | 330 | Neurons, hypodermis, intestine, pharynx (Jiang et al., 2001) | Y | Y |
| hlh-16 | 514 | Head neurons (Bertrand et al., 2011) | Y | N |
| ceh-40 | 965 | Dopaminergic neurons (Sarov et al., 2012) | Y | Y |
| ceh-10 | 1172 | Neurons, seam cells (Reece-Hoyes et al., 2007) | Y | Y |
| ahr-1 | 1387 | ALM and RME neurons (Huang et al., 2004) | Y | N |
| mdl-1 | 2000 | Neurons, body wall, pharynx (Reece-Hoyes et al., 2007) | Y | Y |
| egl-43 | 2001 | Neurons, gonad (Hwang et al., 2007) | Y | N |
| ceh-20 | 2015 | Neurons, seam cells, vulva (Reece-Hoyes et al., 2007) | Y | Y |
| ceh-43 | 2096 | Neurons, anterior hypodermis (Reece-Hoyes et al., 2007) | Y | Y |
| daf-7 | 2524 | Nead neurons, coelemocytes, pharynx (Klabonski et al., 2016) | Y | Y |
| lin-11 | 2857 | Neurons, uterus, vulva, head muscle (Gupta et al., 2003) | Y | Y |
| egl-46 | 4477 | Neurons (Wu et al., 2001) | N | N |
| nhr-67 | 5545 | Neurons, excretory cell, rectal valve cell, vulva (Fernandes and Sternberg, 2007) | Y | Y |
-
Y, yes; N, no.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Caenorhabditis elegans) | aha-1p | wormbase.org | WBGene00000095 | |
| Genetic reagent (C. elegans) | hlh-16p | wormbase.org | WBGene00001960 | |
| Genetic reagent (C. elegans) | ceh-40p | wormbase.org | WBGene00000461 | |
| Genetic reagent (C. elegans) | ceh-10p | wormbase.org | WBGene00000435 | |
| Genetic reagent (C. elegans) | ahr-1p | wormbase.org | WBGene00000096 | |
| Genetic reagent (C. elegans) | mdl-1p | wormbase.org | WBGene00003163 | |
| Genetic reagent (C. elegans) | egl-43p | wormbase.org | WBGene00001207 | |
| Genetic reagent (C. elegans) | ceh-20p | wormbase.org | WBGene00000443 | |
| Genetic reagent (C. elegans) | ceh-43p | wormbase.org | WBGene00000463 | |
| Genetic reagent (C. elegans) | daf-7p | wormbase.org | WBGene00000903 | |
| Genetic reagent (C. elegans) | lin-11p | wormbase.org | WBGene00003000 | |
| Genetic reagent (C. elegans) | egl-46p | wormbase.org | WBGene00001210 | |
| Genetic reagent (C. elegans) | nhr-67p | wormbase.org | WBGene00003657 | |
| Strain, strain background (C. elegans) | N2 | Caenorhabditis Genetics Center | ||
| Strain, strain background (C. elegans) | N2-PD1073 | doi:10.17912/micropub.biology.000518 | Available from the Caenorhabditis Intervention Testing Program- upon request (https://citp.squarespace.com/)- | |
| Strain, strain background (C. elegans) | PX740 | This paper | N2-PD1073 fxIs47 [rsp-0p:: 5′ ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3′ ΔHygR::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | GT331 | This paper | aSi9[lox2272 Cbr-unc-119(+) lox2272+loxP 3′3′ ΔHygR +3′ ΔmScarlet-I::PEST]; unc-119(ed3) | |
| Strain, strain background (C. elegans) | GT332 | This paper | aSi10[lox2272 Cbr-unc-119(+) lox2272+loxP 3′ ΔHygR +3′ ΔmScarlet-I]; unc-119(ed3) | |
| Strain, strain background (C. elegans) | GT336 | This paper | aSi12[lox2272 rps-0p::HygR+hsp−16.41p::Cre::tbb-2 3′UTR+sqt-1(e1350) lox2272+loxN 3′ ΔCbr-unc-119(+)::tjp2a_guide:: 3′ ΔmNeonGreen::PEST::egl-13nls::tbb-2 3′UTR] aSi9[lox2272 Cbr-unc-119(+) lox2272+loxP 3′ΔHygR::tjp2a guide::3′ΔmScarlet-I::PEST::egl-13nls::tbb-2 3′UTR] II; unc-119(ed3) III | |
| Strain, strain background (C. elegans) | GT337 | This paper | aSi13[lox2272+loxN 3' ΔCbr-unc-119(+)+3' ΔmNeonGreen::PEST] aSi14[lox2272+loxP 3′ ΔHygR +3′ ΔmScarlet-I::PEST]; unc-119(ed3), | |
| Strain, strain background (C. elegans) | QL74 | Gift from QueeLim Ch’ng | oxEx1578 [eft-3p::GFP+Cbr-unc-119(+)] 6x outcross EG4322 | |
| Strain, strain background (C. elegans) | PX786 | This paper | fxEx23 [TARDIS #5 5′ ΔHygR::Intron5'::Read1::NNNCNNTNTNANNNN::Read2::Intron3':: 3' ΔHygR (89 Unique Sequences) hsp-16.41p::piOptCas9::tbb-2 34' UTR+rsp-27p::NeoR::unc-54 3' UTR+U6p:: GCGAAGTGACGGTAGACCGT]; fxSi47[ rsp-0p:: 5' ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3' ΔHygR::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | PX816 | This paper | fxEx25 [TARDIS #1 5' ΔHygR::Intron5'::Read1::NNNCNNTNTNANNNN::Read2::Intron3':: 3' ΔHygR (1,319 Unique Sequences) hsp-16.41p::piOptCas9::tbb-2 34' UTR+rsp-27p::NeoR::unc-54 3' UTR+U6p:: GCGAAGTGACGGTAGACCGT]; fxSi47[ rsp-0p:: 5' ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3' ΔHygR::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | PX817 | This paper | fxEx26 [TARDIS #2 5' ΔHygR::Intron5'::Read1::NNNCNNTNTNANNNN::Read2::Intron3':: 3' ΔHygR (3,001 Unique Sequences) hsp-16.41p::piOptCas9::tbb-2 34' UTR+rsp-27p::NeoR::unc-54 3' UTR+U6p:: GCGAAGTGACGGTAGACCGT]; fxSi47[ rsp-0p:: 5' ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3' ΔHygR::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | PX818 profile 1 | This paper | fxEx27 [TARDIS #3 5' ΔHygR::Intron5'::Read1::NNNCNNTNTNANNNN::Read2::Intron3':: 3' ΔHygR (91 Unique Sequences) hsp-16.41p::piOptCas9::tbb-2 34' UTR+rsp-27p::NeoR::unc-54 3' UTR+U6p:: GCGAAGTGACGGTAGACCGT]; fxSi47[ rsp-0p:: 5' ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3' ΔHygR::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | PX818 profile 2 | This paper | fxEx28 [TARDIS #4 5' ΔHygR::Intron5'::Read1::NNNCNNTNTNANNNN::Read2::Intron3':: 3' ΔHygR (204 Unique Sequences) hsp-16.41p::piOptCas9::tbb-2 34' UTR+rsp-27p::NeoR::unc-54 3' UTR+U6p:: GCGAAGTGACGGTAGACCGT]; fxSi47[ rsp-0p:: 5' ΔHygR:: GCGAAGTGACGGTAGACCGT:: 3' ΔHygR′::unc-54 3′::loxP] | |
| Strain, strain background (C. elegans) | PX819 | This paper | N2 fxEx24 [(rps-0p:: 5′ ∆HygR+loxP + aha-1p::SV40 NLS:: 5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + ahr-1p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + ceh-10-1p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + ceh-20p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + ceh-40p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: ∆HygR+loxP + ceh-43p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + daf-7p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: ∆HygR+loxP + egl-43p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + hlh-16p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + lin-11p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + mdl-1p::SV40 NLS::5′ ∆mScarlet-I) + (rps-0p:: 5′ ∆HygR+loxP + nhr-67p::SV40 NLS::5′ ∆mScarlet-I)+hsp−16.41p::piOptCas9::tbb-2 34′ UTR+prsp-27::NeoR::unc-54 3′ UTR]; aSi10[lox2272+Cbr-unc-119(+)+lox2272+loxP + 5′ ∆HygR::unc-54 3' UTR+5′ ∆mScarlet-I::egl-13 NLS::tbb-2 3' UTR, II:8420157]; unc-119(ed3) III | |
| Strain, strain background (C. elegans) | EG4322 | doi.org/10.1038ng.248; Caenorhabditis Genetics Center | ||
| Strain, strain background (Escherichia coli) | PXKR1 | This paper | NA22 transformed with pUC19 | |
| Recombinant DNA reagent | Plasmid pDSP15 | This paper | 193853 (Addgene) | 5′ ΔHygR::loxP::MCS::5′ Δ mScarlet-I |
| Recombinant DNA reagent | Plasmid pDSP16 | This paper | 193854 (Addgene) | 5′ ΔCbr-unc-119(+)::loxN::MCS::5′ Δ 5′mNeonGreen |
| Recombinant DNA reagent | Plasmid pMS84 | This paper | 193852 (Addgene) | U6p::GGACAGTCCTGCCGAGGTGG |
| Recombinant DNA reagent | Plasmid pZCS36 | This paper | 193048 (Addgene) | hsp16.41p::Cas9(dpiRNA)::tbb-2 ′3UTR |
| Recombinant DNA reagent | Plasmid pZCS38 | This paper | 193049 (Addgene) | rsp-27p::NeoR::unc-54 3′ UTR |
| Recombinant DNA reagent | Plasmid pZCS41 | This paper | 193050 (Addgene) | U6p::GCGAAGTGACGGTAGACCGT |
| Sequence-based reagent | ZCS422 | This paper | Design and construction of barcode donor library | |
| Commercial assay or kit | DNA Clean and Concentrator | Zymo Research | Cat# D4004 | |
| Commercial assay or kit | Genomic DNA Clean and Concentrator | Zymo Research | Cat# D4011 | |
| Commercial assay or kit | Zymoclean Gel DNA Recovery Kit | Zymo Research | Cat# D4008 | |
| Commercial assay or kit | Zyppy Plasmid Miniprep Kit | Zymo Research | Cat# D4019 | |
| Software, algorithm | Cutadept | doi.org/10.14806/ej.17.1.200 | Version 4.1 | |
| Software, algorithm | AmpUMI | doi.org/10.1093/bioinformatics/bty264 | Version 1.2 | |
| Software, algorithm | Starcode | doi.org/10.1093/bioinformatics/btv053 | Version 1.4 | |
| Software, algorithm | Google colab | colab.research.google.com | ||
| Software, algorithm | Python (version) | Guido van Rossum, 1991 | Version 3.7.13 | |
| Software, algorithm | Juypter Notebook (IPython) | doi:10.3233/978-1-61499-649-1-87 | Version 7.9.0 | |
| Software, algorithm | matplotlib | doi:10.5281/zenodo.3898017 | Version 3.7.13 | |
| Software, algorithm | Fiji | imagej.net/software/fiji/ | Version 2.9.011.53t | |
| Chemical compound, drug | G-418 | GoldBio (CAS number 108321-42-2) | Cat# G-418-5 | |
| Chemical compound, drug | Hygromycin B | GoldBio (CAS number 31282-04-9) | Cat# H-270-10-1 |
Additional files
-
Supplementary file 1
Table of the plasmids used.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp1-v1.xlsx
-
Supplementary file 2
Table of the primers used for cloning.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp2-v1.xlsx
-
Supplementary file 3
Table of the primers used for strain confirmation by Sanger sequencing.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp3-v1.xlsx
-
Supplementary file 4
Table of the primers and ultramer used to create the barcode donor homology.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp4-v1.xlsx
-
Supplementary file 5
Table of the primers used for Illumina sequencing.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp5-v1.xlsx
-
Supplementary file 6
Table of the primers used for promoter identification.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp6-v1.xlsx
-
Supplementary file 7
Complete genotypes for the barcode TARDIS arrays presented in the study.
- https://cdn.elifesciences.org/articles/84831/elife-84831-supp7-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84831/elife-84831-mdarchecklist1-v1.docx





