The dark kinase STK32A regulates hair cell planar polarity opposite of EMX2 in the developing mouse inner ear
Figures
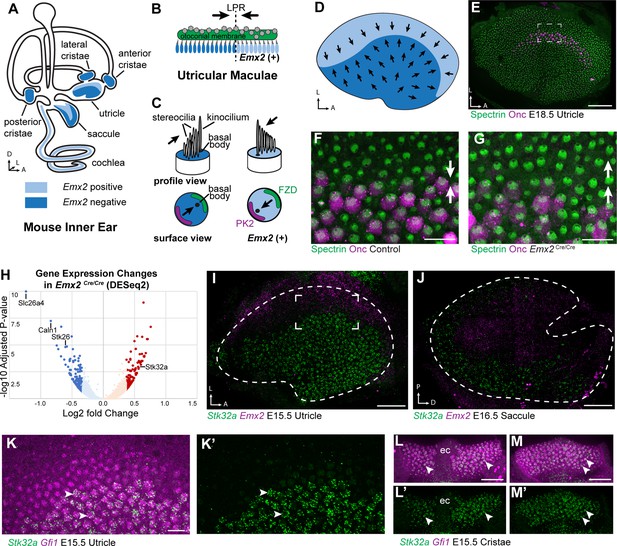
Transcriptome analysis of Emx2 mutant utricles identifies Stk32a as a candidate planar polarity effector.
(A) The sensory epithelia (blue shading) of the mouse inner ear contains hair cells that can be divided into those that express EMX2 and those that do not. (B) The utricle contains two groups of hair cells patterned about the LPR and oriented to detect movements of the otoconial membrane directed in opposite directions (arrows). (C) Utricular hair cells with opposite stereociliary bundle orientations (arrows) viewed in profile or from a top-down surface view. The cuticular plate (blue) can be used to determine bundle orientation based upon the position of the basal body. The Core PCP proteins PK2 and FZD are similarly distributed in hair cells with either bundle orientation. (D) The border of EMX2 expression distinguishes hair cells with opposite bundle orientations (arrows). (E) E18.5 mouse utricle labeled for βII-Spectrin to mark the hair cell cuticular plates and Oncomodulin to mark the striolar region. (F) Higher magnification of the framed region from ‘E’ illustrating the orientation (arrows) of the two hair cell groups. (G) In Emx2 Cre/Cre mutants the hair cells are oriented in a single direction. (H) Gene expression changes in Emx2 Cre/Cre utricles relative to littermate control determined by bulk RNAseq analysis of micro-dissected tissue. Select upregulated and downregulated genes evaluated in subsequent figures are annotated. (I) Fluorescent ISH showing the complementary patterns of Emx2 and Stk32a mRNA expression in E15.5 utricle or (J) E16.5 saccule. Dashed lines mark the boundary of the sensory epithelia determined by Gfi1 expression in a third channel. (K,K’) Higher magnification of the framed region in ‘I’ showing the cellular distribution of Stk32a mRNA and overlap with Gfi1 in medial hair cells. (L,L’) Stk32a similarly colocalizes with Gfi1 in the anterior cristae and (M,M’) horizontal cristae. Arrowheads highlight individual hair cells. (ec) Eminentia Cruciatum. Scale bars: E,I,J,L,M (50 µm); F,G,K (20 µm).
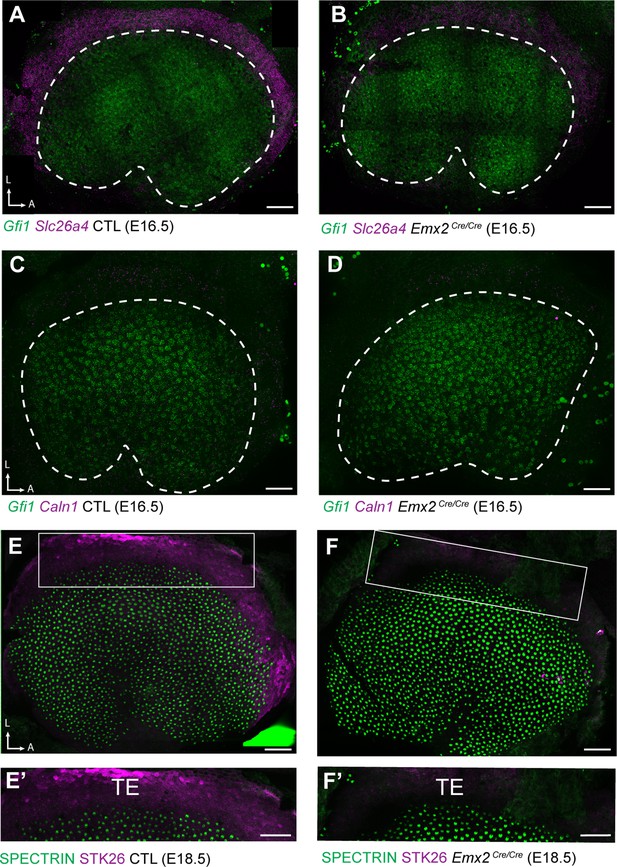
Down-regulated genes are enriched in TE.
(A) fISH for Slc26a4 in control utricles at E16.5 reveals expression in the transitional epithelia (TE) region located adjacent to utricular hair cells expressing Gfi1. (B) Slc26a4 fISH fluorescence intensity is reduced in the TE region of Emx2 Cre/Cre mutant utricle consistent with RNAseq. (C) Caln1 mRNA is similarly enriched in the TE and (D) reduced in Emx2 Cre/Cre mutant utricles. Dashed lines demarcate the boundary of the sensory epithelia based upon Gfi1 expression. (E) Immunolabeling of STK26 at E18.5 marks the TE adjacent to βII-Spectrin labeled hair cells in the sensory epithelia. (E’) High magnification image of the boxed region in ‘E’. (F) STK26 immunolabeling is lost from the TE of Emx2 Cre/Cre mutants. (F’) High-magnification image of the boxed region in ‘F’. Scale bars: 50 µm.
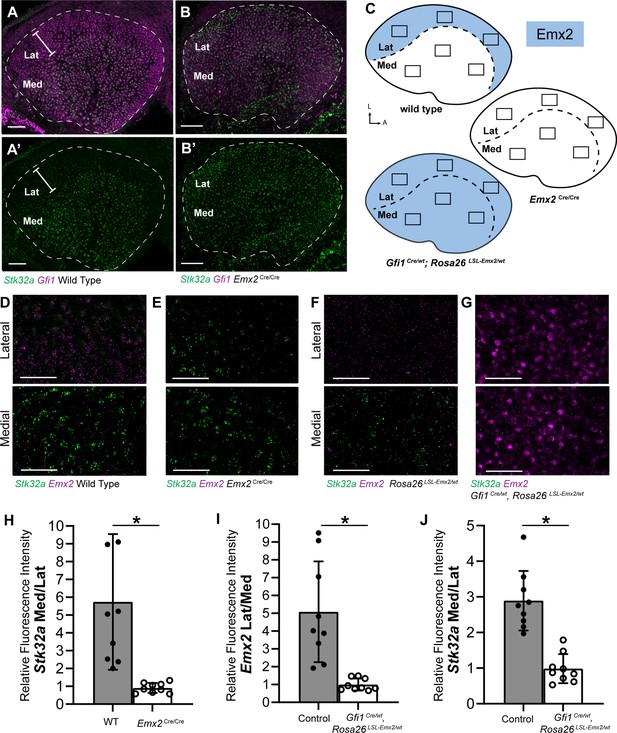
Stk32a expression is inversely correlated with Emx2.
(A,A’) Stk32a mRNA is restricted to the medial region (Med) of the mouse utricle and expands in Emx2Cre/Cre mutants (B,B’) to overlap with the hair cell marker Gfi1 throughout the sensory epithelium. (C) Emx2 distribution in wild type tissues, Emx2 Cre/Cre mutants or following transgenic overexpression (Gfi1 Cre/wt; ROSA Emx2/wt). Boxed regions indicate relative positions of analysis fields used for fluorescence intensity measures. (D,E) Representative images used to measure Emx2 and Stk32a wISH fluorescence in control and Emx2 Cre/Cre mutant utricles. (F,G) Representative images used to measure Emx2 and Stk32a wISH fluorescence from Cre-negative controls and Gfi1-Cre, Rosa26 LSL-Emx2/wt transgenic mice overexpressing Emx2 in hair cells. (H) Relative fluorescence for Stk32a in medial (Med) vs. lateral (Lat) regions following Emx2 gene deletion. (I) Relative fluorescence for Emx2 in lateral vs. medial and (J) Stk32a in medial vs. lateral regions following Emx2 overexpression. Control mice used for quantification are Cre-negative. Pairwise comparisons of relative fluorescence intensity were evaluated by Student’s test (* p<0.002) and error bars indicate SD. Scale bars: A,B (50 µm); D-G (25 µm).
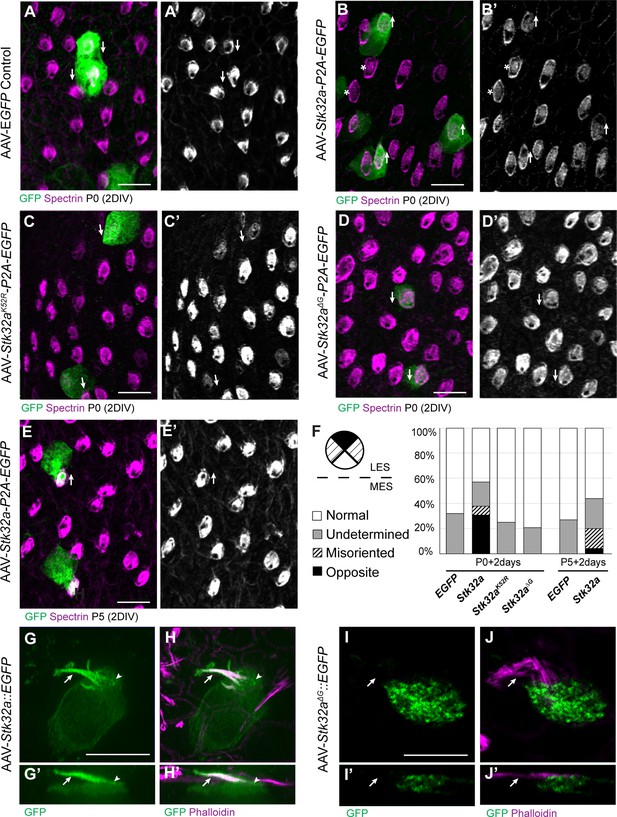
STK32A is sufficient to reorient stereociliary bundles in the Emx2 domain.
(A,A’) Hair cells from the lateral region transduced with AAV vectors expressing EGFP at P0 develop a cuticular plate with the fonticulus positioned towards the LPR when evaluated by βII-Spectrin labeling. Arrows illustrate orientation of transduced cells. (B,B’) Hair cells from the lateral region expressing STK32A and EGFP frequently developed medial bundle orientations as revealed by the position of the fonticulus. Arrows illustrate orientation of transduced cells and asterisks mark examples of non-transduced cells with undetermined bundle orientation. (C,C’) Hair cells from the lateral region expressing the kinase mutant STK32A K52R. (D,D’) Hair cells from the lateral region expressing the STK32A Δ2G mutant lacking the predicted N-myristoyl glycine. (E,E’) Medial bundle orientations are less frequent in lateral hair cells transduced at P5. (F) Frequency of reoriented or misoriented stereociliary bundles for AAV transduced hair cells in the lateral extrastriolar of utricular explants cultured from P0 and P5 mice. Cells with a fonticulus located in the apical cell surface quadrant closest to the LPR were considered normal while cells with a fonticulus in the quadrant furthest from the LPR were classified as Opposite. Cells with a poorly resolved fonticulus were classified as Undetermined while all other orientations were considered Misoriented for this quantification. For AAV-Stk32a-P2A-EGFP at P0 (N=433 cells from 5 utricles), AAV-Stk32a-P2A-EGFP at P5 (N=109 cells from 4 utricles), AAV-EGFP at P0 (N=167 cells from 3 utricles), AAV-EGFP at P5 (N=165 cells from 3 utricles), AAV-Stk32a Δ2G-P2A-EGFP at P0 (N=106 cells from 4 utricles), AAV-Stk32a (K52R)-P2A-EGFP at P0 (N=108 cells from 4 utricles). (E–G) Hair cells transduced with virus expressing STK32A::EGFP (G,G’) fusion show EGFP immunofluorescence throughout the stereociliary bundle (arrow) that overlaps with phalloidin (H,H’) and at the apical surface (arrowhead). Hair cells transduced with STK32A Δ2G::EGFP lacking N-myristoylation does not colocalize with phalloidin in the apical compartment or bundle (G’,H’,I’,J’) Cellular profiles extracted from image stacks corresponding to G,H,I&J. Scale bars: A-C (10 µm), E (5 µm).
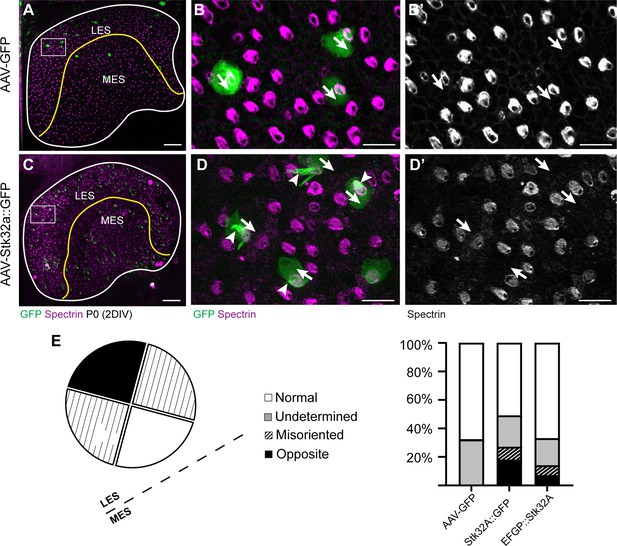
STK32A::EGFP (C-terminal fusion) is functionally active.
(A) P0 Mouse utricle explants cultured in vitro with AAV expressing EGFP results in transduced hair cells on either side of the LPR (yellow line). (B) High-magnification image of boxed region in ‘A’ illustrating individual EGFP-expressing hair cells and βII-SPECTRIN labeling of cuticular plates. (B’) Arrows illustrate orientation of transduced cells. (C) AAV expressing STK32A with a C-terminal EGFP tag (STK32A::GFP) similarly transduces hair cells locate throughout utricle explants. (D) High-magnification image of boxed region in ‘C’ illustrating individual EGFP-expressing hair cells and βII-SPECTRIN labeling of the cuticular plate. STK32A::GFP also appears in the stereociliary bundle (arrowheads). (D’) Arrows illustrate orientation of transduced cells. Similar to untagged STK32A, STK32A::EGFP can produce medial bundle orientations in hair cells located in the lateral extrastriolar region. (E) Quantification of the frequency of bundle reorientation suggests that addition of EGFP reduces STK32A function in this assay, but that this negative effect is greater for N-terminal EGFP::STK32A than the C-terminal EGFP. Scale bars: A, C (50 µm), B, B’, D, D’ (10 µm).
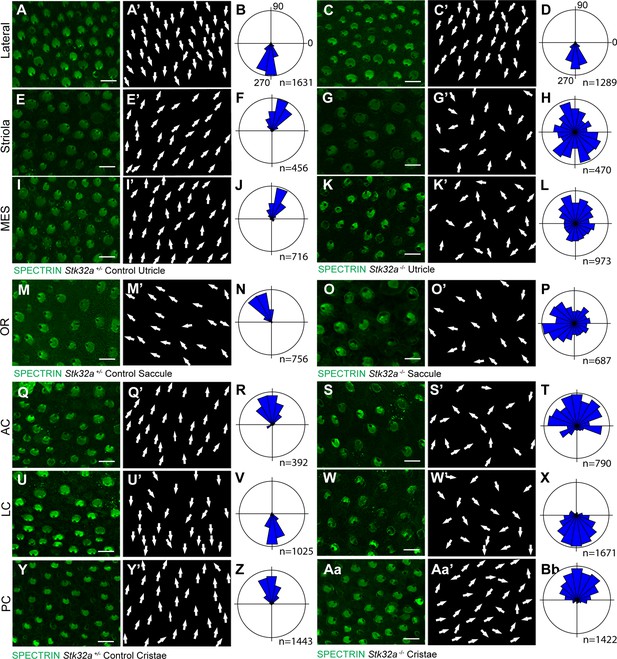
STK32A coordinates stereociliary bundle orientation with the PCP axis.
Stereociliary bundle orientation in inner ear sensory epithelia from P5 heterozygous control and Stk32a -/- mice determined by labeling the cuticular plate with antibodies against βII-SPECTRIN. (A,A’,C,C’) Hair cells in the lateral region of the utricle retained normal bundle orientations in both genotypes. (E,E’,G,G’) Hair cells in the Stk32a -/- striolar region appear randomly oriented relative to controls. (I,I’,K,K’) Hair cells in the Stk32a -/- medial extrastriolar region (MES) appear randomly oriented relative to controls. (M,M’,O,O’) Hair cells in the outer region of the Stk32a -/- saccule appear disorganized relative to controls. (Q,Q’,U,U’,Y,Y’) Hair cells in cristae of controls are tightly aligned with the associated semi-circular canal while (S,S’,W,W’,Aa,Aa’) Stk32a -/- cristae contain disorganized hair cell bundles. Cumulative graphs of individual bundle orientations for each sensory epithelia pooled from multiple littermate controls (B,F,J,N,R,V,Z) or Stk32a -/- (D,H,L,P,T,X,Bb) (N=3–4 sensory organs for each). Summary of analysis field locations is illustrated in Figure 4—figure supplement 1. (MES) medial extrastriolar, (OR) outer region, (AC) anterior cristae, (LC) lateral cristae, (PC) posterior cristae. Scale bars: 10 µm.
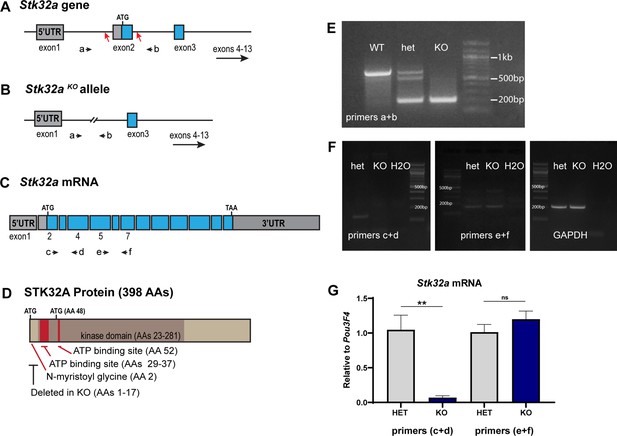
Stk32a gene targeting strategy and validation.
(A) The Stk32a gene was edited using CRISPR/Cas9 and guide RNAs targeting CRISPR cut sites (red arrows) flanking exon 2, which contains the translational start. (B,E) Mice in which non-homologous end-joining repair of these two cuts and deleted exon2 were identified by PCR using primer pairs (a+b) flanking exon 2. (C,F) Stk32a expression was evaluated by RT-PCR using primer pairs that amplified portions of the mRNA encoded by exon2 or downstream exons that were not impacted by the Stk32a -/- deletion, and suggested that mutant mRNA persisted. (G) Quantitative RT-PCR measures of Stk32a mRNA relative to the hair cell transcription factor Pou3F4 confirmed that mutant mRNA was not eliminated by nonsense mediated decay. Pairwise comparisons of gene expression were evaluated by Student’s test (** p<0.001) and error bars indicate SEM. (D) The N-myristoyl Glycine, STK32A protein kinase domain and essential ATP binding sites identified by sequence homology to related kinases (UniProt). These regions are illustrated relative to the translational start (ATG) and an alternative ATG downstream of exon2. Any mutant protein translated from this or subsequent ATG codons would lack an intact ATP binding domain and would therefore be non-functional as a kinase, and further would lack the N-myristoyl-glycine that is also necessary for STK32A to reorient lateral hair cells in explant cultures.
-
Figure 4—figure supplement 1—source data 1
DNA electrophoresis of PCR products.
(A) Original and (A’) annotated PCR gel demonstrating the Stk32a gene deletion generated in the Stk32a KO line. (B) Original and (B’) annotated RT-PCR gel demonstrating the absence of Stk32a coding sequence exon 2 in the Stk32a KO line and GAPDH control reactions. (C) Original and (C’) RT-PCR gel demonstrating the presence of 3’ Stk32a coding sequence exons in the Stk32a KO line. Dashed lines indicated portions of each gel that were cropped and presented in (Figure 4—figure supplement 1). Remaining lanes contain reactions that either failed or were not pertinent to that figure.
- https://cdn.elifesciences.org/articles/84910/elife-84910-fig4-figsupp1-data1-v2.pdf
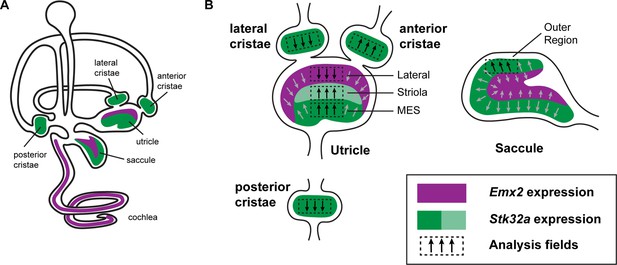
Emx2 and Stk32a expression domains and planar polarity analysis fields.
(A) Within the sensory epithelia of the mouse inner ear there are complementary and non-overlapping domains of Emx2 and Stk32a expression. Gene expression outside of the sensory epithelia is not included. (B) Dashed boxes indicate the position of each analysis field used for stereociliary bundle orientation measures in Figure 3. Black arrows indicate general orientation within these analysis fields for hair cells from wild type mice. For the developing utricle, three separate domains were included that flank the LPR. These are the lateral domain which expresses Emx2, and the striola and medial extrastriolar domain (MES) that express Stk32a.
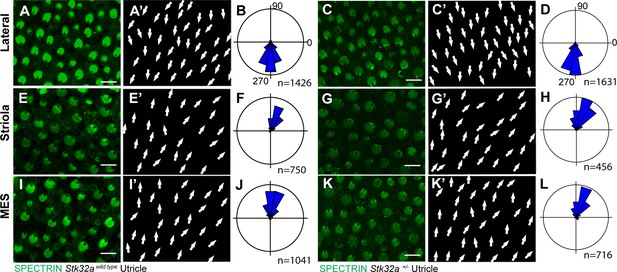
Stk32a+/-mice do not display planar polarity phenotypes.
(A) Stereociliary bundle orientation in inner ear sensory epithelia from P5 wild type and Stk32a +/- mice determined by labeling the cuticular plate with antibodies against βII-SPECTRIN. (A,A’,C,C’) Hair cells in the lateral region of the utricle have similar organization and bundle orientations for both genotypes. (E,E’,G,G’) Hair cells in the region of the utricle have similar organization and bundle orientations for both genotypes. (I,I’,K,K’) Hair cells in the medial extrastriolar region (MES) of the utricle have similar organization and bundle orientations for both genotypes. For this comparison Stk32a +/- images and histograms are reproduced from Figure 4.
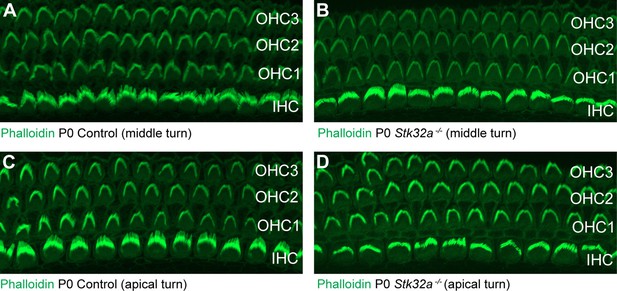
Auditory hair cells are not impacted by Stk32a gene deletion.
(A,C) Phalloidin labeling of auditory hair cells from the middle (A) and apical (B) turns of the P0 mouse cochlea. (B,D) Phalloidin labeling of auditory hair cells from Stk32a -/- cochlea reveals no changes in the morphology or planar polarity of the stereociliary bundle in either the middle (B) or apical (D) turns. (IHC) Inner hair cell, (OHC1) Outer hair cell row 1, (OHC2) Outer hair cell row 2, (OHC3) Outer hair cell row 3.
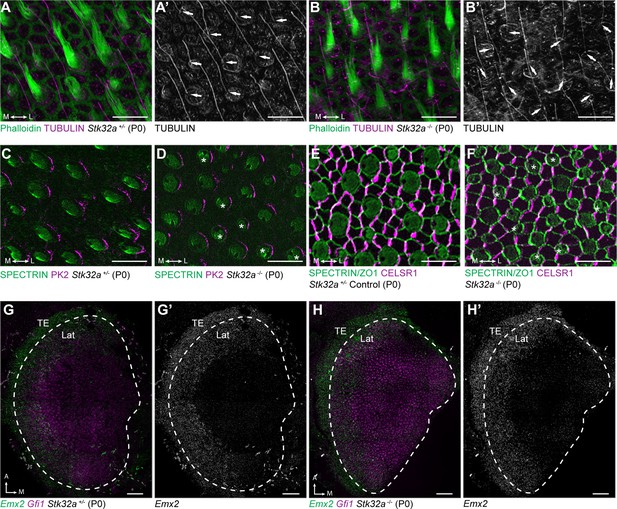
Stereociliary bundle polarization, PCP and LPR patterning remains intact in Stk32a mutants.
(A) P0 heterozygous control and (B) Stk32a -/- stereociliary bundles immunolabeled with acetylated-tubulin antibodies to mark the kinocilium and phalloidin to label the stereocilia. Stk32a -/- hair cells from intact polarized bundles despite being misoriented. Arrows indicated bundle orientation. (C,D) PK2 is asymmetrically distributed along one side of vestibular hair cells from the striolar region of control (C) and Stk32a -/- (D) utricles. Asterisks mark examples of misoriented cells where the fonticulus is not correctly positioned opposite of PK2 in the Stk32a -/-. (E,F) CELSR1 immunolabeling at junctions between supporting cells reveals the orientation of the PCP polarity axis in heterozygous control (E) and Stk32a -/- (F) utricles. Combined labeling of βII-SPECTRIN and ZO-1 reveals hair cell orientation and junctions between supporting cells. Asterisks mark examples of misoriented hair cells in Stk32a -/-. (G,G’) fISH for Emx2 and Gfi1 demonstrating Emx2 expression in the lateral utricle and adjacent transitional epithelia (TE) of littermate controls. Dashed lines demarcate the boundaries of Gfi1-expressing hair cells. (H,H’) In Stk32a -/- utricles, the Emx2 expression domain is not changed relative to Gfi1. Scale bars:A-F (10 µm); G,H (50 µm).
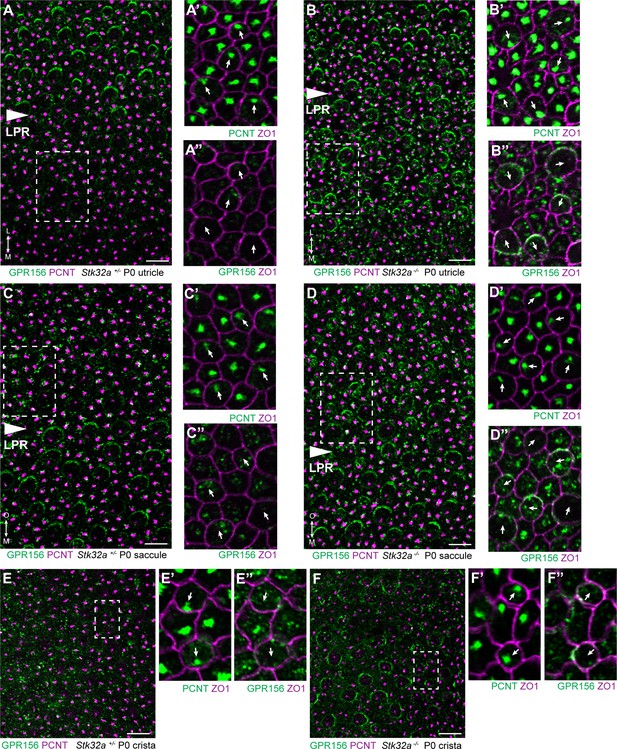
STK32A regulates GPR156 distribution at the apical cell surface.
(A) GPR156 is asymmetrically distributed at apical cell boundaries of hair cells in the lateral region of the utricle along one side of the LPR. Pericentrin (PCNT) shows the position of the basal body beneath cilia in hair cells and supporting cells. (A’) Higher magnification image of the boxed region from ‘A’ taken from the medial side of the LPR. Pericentrin and ZO-1 labeling reveals the position of the basal body and hence orientation of the stereociliary bundle (arrows). (A’’) The distribution of GPR156 relative to ZO1 in the boxed region from ‘A’. (B-B’’) In Stk32a -/- utricles, GPR156 is redistributed to the apical boundaries of hair cells on the medial side of the LPR, although the uniform asymmetric distribution is lost. (C-C’’) GPR156 is asymmetrically distributed at apical cell boundaries of hair cells in the inner region of the saccule along one side of the LPR. (D-D’’) GPR156 is similarly redistributed and can be detected at the apical boundaries of hair cells on either side of the LPR in the Stk32a -/- saccule. For these experiments the position of the LPR was determined based upon the organization of stereociliary bundles which are aligned in the Emx2-positive region and misoriented in the Emx2-negative region in Stk32a -/-. (E-E’’) In littermate control cristae GPR156 signal is visible but not at apical cell boundaries. (F-F’’) In Stk32a -/- cristae, GPR156 is redistributed to the apical cell boundaries, although similar to the utricle and saccule GPR156 distribution is not uniformely planar polarized. Scale bars: 10 µm.
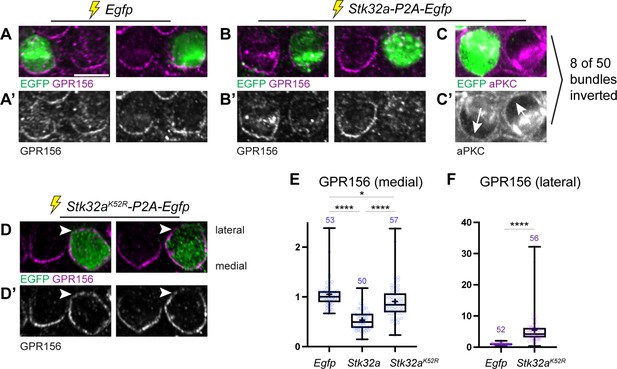
Ectopic expression of STK32A in auditory hair cells disrupts GPR156 localization.
(A,A’) Cochlea electroporated at E13.5 and grown in culture develop auditory hair cells with distinct crescents of GPR156 enriched at their medial cell boundaries. Two examples of an EGFP-positive electroporated cell with a non-electroporated neighbor are provided. (B,B’) When electroporated with Stk32a-P2A-EGFP, hair cells expressing the EGFP reporter have a stark reduction in GRP156 at their apical cell boundaries. (C,C’) A minority of cells expressing STK32A (8/50, 16%) had reversed planar polarity as revealed by the distribution of the intrinsic polarity marker aPKC. Arrows indicated bundle orientations. (D,D’) GPR156 distribution at the medial cell boundary of hair cells expressing the predicted kinase-dead STK32A K52R mutant was only mildly impacted while GPR156 distribution at the lateral boundary of these cells increased (arrowheads). (E) The intensity of GPR156 immunolabel fluorescence at medial cell boundaries in electroporated relative to neighboring cells is significantly reduced in cells that ectopically express STK32A and only modestly impacted in cells expressing STK32A K52R. Pairwise comparisons of relative fluorescence intensity were evaluated by one-way Anova (**** p<0.0001, * p=0.0127). (F) The intensity of GPR156 immunolabel fluorescence at lateral cell boundaries in electroporated relative to neighboring cells is significantly increased in cells that ectopically express STK32A K52R. Pairwise comparisons of relative fluorescence intensity were evaluated using the Mann-Whitney test (**** p<0.0001). Gpr156 data is graphed as 25-75% boxplots where exterior lines show minimum and maximum values, the middle line the median and + the mean. Scale bars: 5 µm.
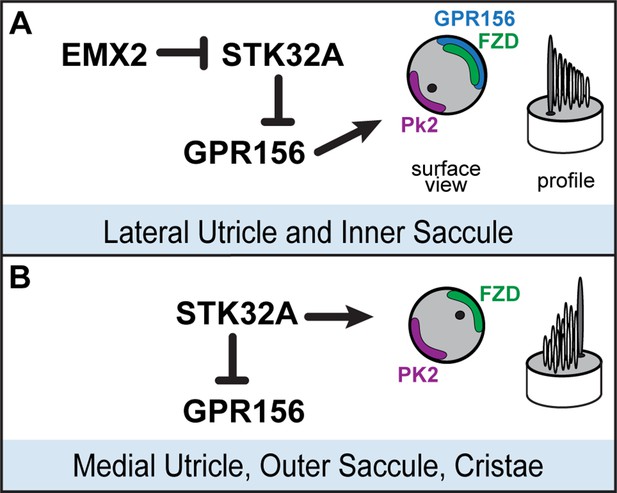
Model for patterning hair cells about the LPR.
(A) In the lateral region of the utricle and inner region of the saccule, GPR156 functions to orient the stereociliary bundle relative to the PCP axis so that the basal body and kinocilium are positioned adjacent to PK2. Repression of STK32A transcription by EMX2 enables GPR156 function and establishes the position of the LPR. (B) In the medial utricle, outer region of the saccule and the semi-circular canal cristae, STK32A orients the stereociliary bundle so that the basal body and kinocilium are positioned adjacent to FZD. Although the direct substrates of STK32A in these regions are not known, STK32A-dependent regulation of GPR156 at apical cell boundaries prevents orientation reversal and reinforces the position of the LPR.
Additional files
-
Supplementary file 1
Down-regulated genes in the Emx2 Cre/Cre utricle.
Summary of genes that are down-regulated in the absence of Emx2. Sequencing results were analyzed using Bioconductor DESeq as well as post hoc-analysis using Swish, and Benjamini-Hirshberg false discover rate was used to normalize data across all transcripts and produce adjusted p-values based on log2-fold changes between Emx2 Cre/Cre and WT controls. Only genes with adjusted P-values <0.05 are listed. Additional data for genes highlighted with blue shading are presented in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/84910/elife-84910-supp1-v2.xlsx
-
Supplementary file 2
Up-regulated genes in the Emx2 Cre/Cre utricle.
Summary of genes that are up-regulated in the absence of Emx2. Sequencing results were analyzed using Bioconductor DESeq as well as post hoc-analysis using Swish, and Benjamini-Hirshberg false discover rate was used to normalize data across all transcripts and produce adjusted p-values based on log2-fold changes between Emx2 Cre/Cre and WT controls. Only genes with adjusted P-values <0.05 are listed. Red shading highlights Stk32a.
- https://cdn.elifesciences.org/articles/84910/elife-84910-supp2-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/84910/elife-84910-mdarchecklist1-v2.docx





