Prolonged T-cell activation and long COVID symptoms independently associate with severe COVID-19 at 3 months
Figures
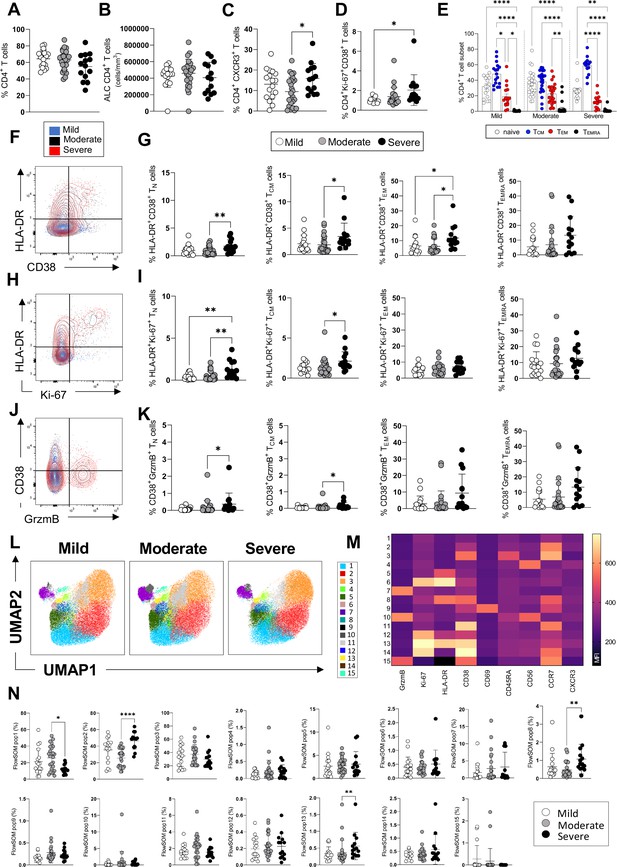
CD4+ T-cell profiles in convalescent coronavirus disease-19 (COVID-19) patients at 3 months post admission.
(A–D) Percentage of CD4+ T-cells within the CD3+ gate (A), absolute number of CD4+ T-cells (cells/mm3) (B), and percentages of CD4+ T-cells expressing CXCR3 (C) and co-expressing Ki67/CD38 (D) are shown in mild, moderate, and severe patients. (E) Percentages of naïve (CCR7+ CD45RA+), T central memory (TCM, CCR7+ CD45RA-), T effector memory (TEM, CCR7- CD45RA-), and T effector memory RA re-expressing (TEMRA, CCR7- CD45RA+) CD4+ T-cells are shown for patients with mild, moderate, and severe disease. (F) Flow cytometry plot showing a representative staining from a mild, moderate, and severe patient of HLA-DR and CD38 expression in CD4+ TEM cells (overlaid and shown respectively in blue, black, and red). (G) Percentages of activated HLA-DR+CD38+ CD4+ T-cells within naïve, TCM, TEM, and TEMRA cells. (H) Flow cytometry plot with a representative staining from a mild, moderate, and severe patient of HLA-DR and Ki67 expression in CD4+ TEM cells. (I) Percentages of proliferating HLA-DR+ Ki67+ CD4+ T-cells within naïve, TCM, TEM, and TEMRA cells. (J) Flow cytometry plot with a representative staining from a mild, moderate, and severe patient of HLA-DR and granzyme B (GrzmB) expression in CD4+ TEM cells. (K) Percentages of proliferating HLA-DR+ GrzmB+ CD4+ T-cells within naïve, TCM, TEM, and TEMRA cells. (L) Unsupervised uniform manifold approximation and projection (UMAP) analysis showing the FlowSOM clusters in mild (N=17), moderate (N=25), and severe (N=14) patients. Plots are gated on CD4+ T-cells. (M) Heatmap with the expression of each analysed marker within the FlowSOM populations shown as mean fluorescence intensity (MFI). (N) Summary of the percentage of CD4+ T-cells within the indicated FlowSOM populations in mild, moderate, and severe patients. Data in graphs are visualised as mean ± SEM. Statistics are calculated by one-way ANOVA (Kruskal-Wallis test) with Dunn’s correction for multiple testing.
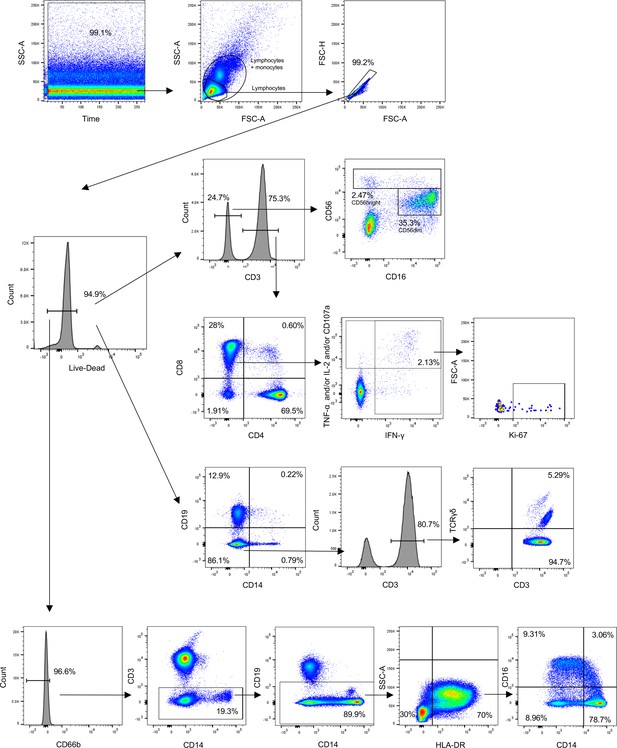
Gating strategy used to identify CD4+, CD8+, and TCR-γδ T-cells, NK cells, and monocytes.

Dynamic changes of immune populations and inflammatory markers in coronavirus disease-19 (COVID-19) patients at acute illness, 3 and 8 months post admission.
(A–F) Lymphocyte (A–C) and neutrophil (D–F) counts during acute illness, at 3 and 8 months post admission in patients with mild (A, D), moderate (B, E), and severe (C, F) disease. (G–L) Albumin (G–I) and CRP (J–L) levels during acute illness, at 3 and 8 months post admission in patients with mild (G, J), moderate (H, K), and severe (I, L) disease. Data from mild (acute: N=17; 3 months: N=17; 8 months: N=10), moderate (acute: N=32; 3 months: N=32; 8 months: N=21), and severe (acute: N=14; 3 months: N=14; 8 months: N=11) patients are indicated with white, grey, and black symbols. Data are shown as a mean ± SD; *p<0.05, **p≤0.01, ***p≤0.001. Statistics were calculated by one-way ANOVA, with Geisser-Greenhouse correction for multiple testing.
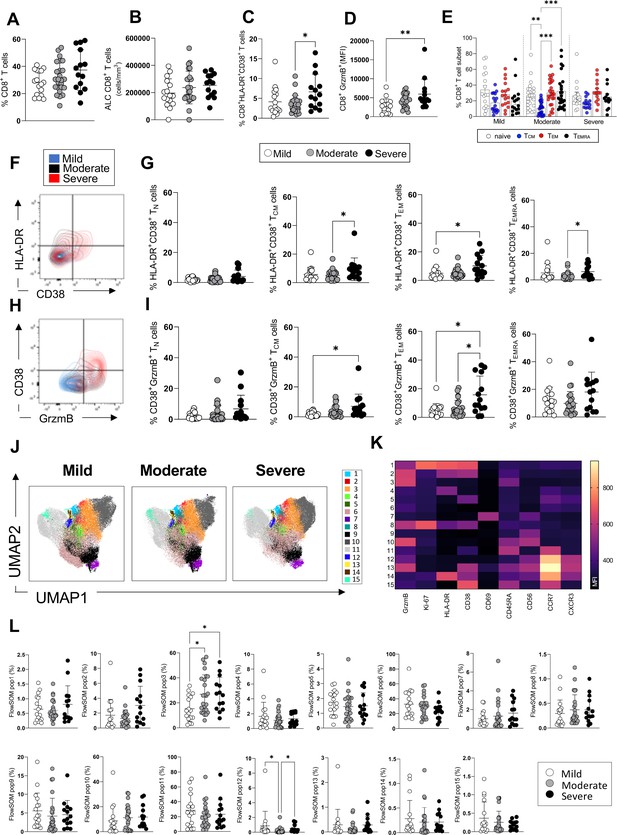
CD8+ T-cell profiles in convalescent coronavirus disease-19 (COVID-19) patients at 3 months post admission.
(A–D) Percentage of CD8+ T-cells within the CD3+ gate (A), absolute number of CD8+ T-cells (cells/mm3) (B), and percentages of CD8+ T-cells co-expressing the activation markers HLA-DR/CD38 (C) or granzyme B (D, shown as mean fluorescence intensity [MFI]) are shown in mild, moderate, and severe patients. (E) Percentages of naïve (CCR7+ CD45RA+), T central memory (TCM, CCR7+ CD45RA-), T effector memory (TEM, CCR7- CD45RA-), and T effector memory RA re-expressing (TEMRA, CCR7- CD45RA+) CD8+ T-cells in patients with mild, moderate, and severe disease. (F) Flow cytometry plot with a representative staining from a mild, moderate, and severe patient (overlaid and shown respectively in blue, black, and red) of HLA-DR and CD38 expression in CD8+ TEM cells. (G) Percentages of activated HLA-DR+ CD38+ CD8+ T-cells within naïve, TCM, TEM, and TEMRA cells. (H) Flow cytometry plot with a representative staining from a mild, moderate, and severe patient of HLA-DR and granzyme B (GrzmB) expression in CD8+ TEM cells. (I) Percentages of proliferating HLA-DR+ GrzmB+ CD8+ T-cells within naïve, TCM, TEM, and TEMRA cells. (J) Unsupervised uniform manifold approximation and projection (UMAP) analysis showing the FlowSOM clusters in mild (N=17), moderate (N=25), and severe (N=14) patients. Plots are gated on CD8+ T-cells. (K) Heatmap with MFI levels for each analysed marker within the FlowSOM populations. (L) Summary of percentage of CD8+ T-cells within the indicated FlowSOM populations in mild, moderate, and severe patients. Data in the graphs are shown as mean ± SEM. Statistics were calculated by one-way ANOVA (Kruskal-Wallis test) with Dunn’s correction for multiple testing.
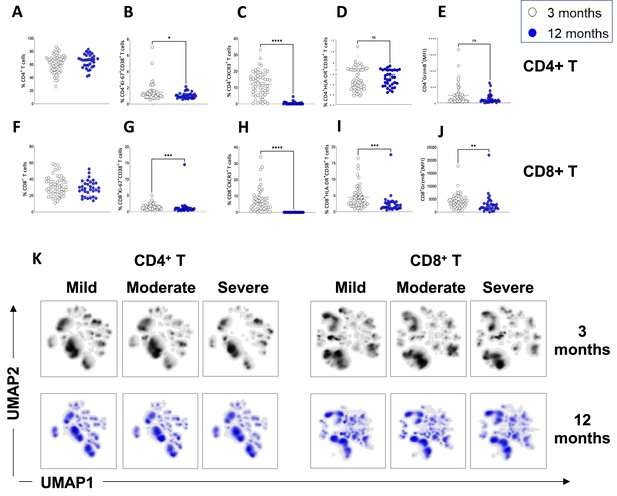
Resolution of T-cell activation at 12 months.
(A–E) CD4+ T-cells in convalescent coronavirus disease-19 (COVID-19) patients at 3 and 12 months. Shown are the percentages of CD4+ T-cells within the CD3+ gate (A) and the percentages of CD4+ T-cells that are Ki67+ CD38+ (B), CXCR3+ (C), HLA-DR+CD38+ (D) and granzyme B+ (shown as mean fluorescence intensity [MFI]) (E).(F–J) CD8+ T-cells in convalescent COVID-19 patients at 3 and 12 months. Shown are the percentages of CD8+ T-cells within the CD3+ gate (F) and the percentages of CD8+ T-cells that are Ki67+ CD38+ (G), CXCR3+ (H), HLA-DR+CD38+ (I), and granzyme B+ (shown as MFI) (J). (K) Unsupervised uniform manifold approximation and projection (UMAP) analysis showing the density plot of cell distribution in mild (N=17), moderate (N=25), and severe (N=14) patients at 3 months and in matched mild (N=8), moderate (N=17), and severe (N=8) patients at 12 months post infection. Data in A–J are shown as mean ± SEM. Statistics in A–J were calculated by Mann-Whitney t-test.
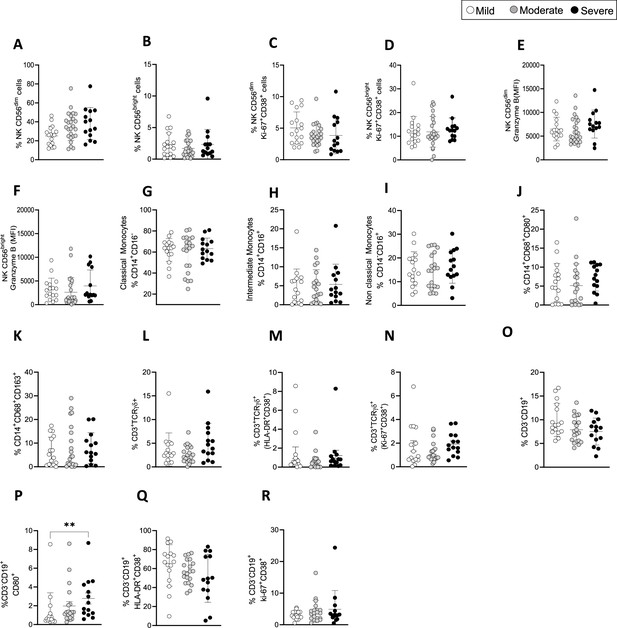
Immune cell populations in coronavirus disease-19 (COVID-19) patients at 3 months post admission.
(A–F) Shown are the percentages of CD56dim (A), CD56bright NK cells (B), Ki67+CD38+ CD56dim (C), and Ki67+CD38+CD56bright NK cells (D). Granzyme B expression on CD56dim (E) and CD56bright (F) NK cells is shown as mean florescence intensity (MFI). (G–K) Shown are the percentages of classical (G: CD14+CD16-), intermediate (H: CD14+CD16+), and non-classical (I: CD14-CD16+) monocytes, activated CD14+CD80+CD86+ (J) and CD14+CD86+CD163+ cells (K). (L–N) TCR-γδ T-cells: frequencies of TCR-γδ T-cells (L), activated HLA-DR+CD38+ (M), and activated/proliferating Ki67+CD38+ TCR-γδ T-cells (N).(O–R) Shown are the percentages of CD3-CD19+ B cells (O), activated CD80+ (P), and HLA-DR+CD38+ B cells (Q) and proliferating Ki67+CD38+ B cells (R). Data are shown as mean ± SD in patients with mild (N=16), moderate (N=21) and severe (N=14) disease. Statistics were by calculated one-way ANOVA test (Kruskal-Wallis test) with Dunn’s correction for multiple testing.
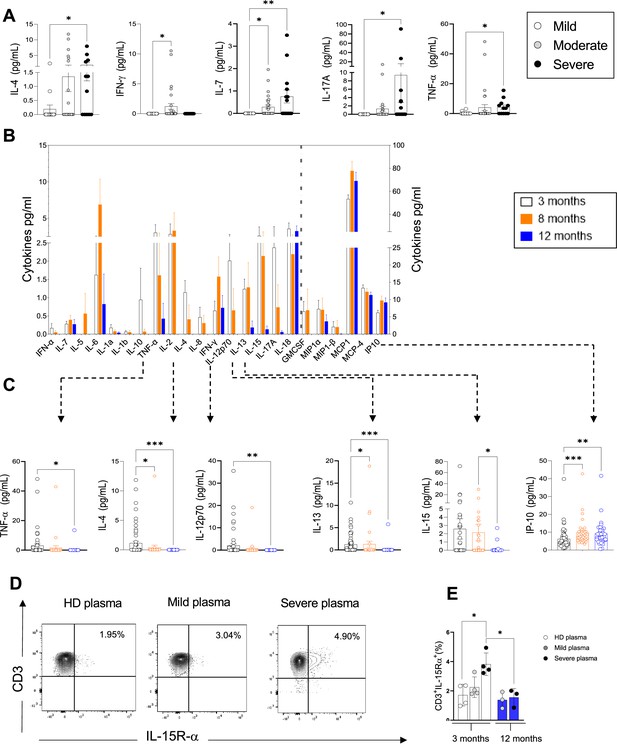
Plasma pro-inflammatory cytokines/chemokines measured at 3, 8, and 12 months.
(A) Plasma cytokines/chemokines measured at 3 months post admission which differed significantly between patients with mild, moderate, and severe disease are shown (N=63: mild: N=17; moderate: N=32; severe: N=14, depicted in white, grey, and black bars, respectively). (B–C) Cytokines/chemokines measured longitudinally in matched samples in patients at 3 (n=63), 8, and 12 months post admission (n=33 samples for each time point) are shown. Data from analytes that differed significantly between time points in B are shown in C for each patient. (D, E) Purified CD3+ T-cells from healthy donors (N=4 for 3 months; N=3 for 12 months) were co-cultured with plasma from 4 healthy donors, 4 mild, and 4 severe patients at 3 months post infection. Shown is IL-15R-α expression in T-cells from a representative donor at 3 months (D) and the average expression of IL-15Rα by T-cells from each peripheral blood mononuclear cell (PBMC) donor after co-culture with plasma from healthy, mild and severe patients, where each data point represents a single patient (E). Statistics were calculated by one-way ANOVA test (Kruskal-Wallis test) with Dunn’s multiple comparison test (A, E) and by ANOVA/repeated-measures one-way ANOVA, mixed-effects analysis with the Geisser-Greenhouse correction, Tukey’s multiple comparison test. (B, C) Data are visualised as mean ± SEM.
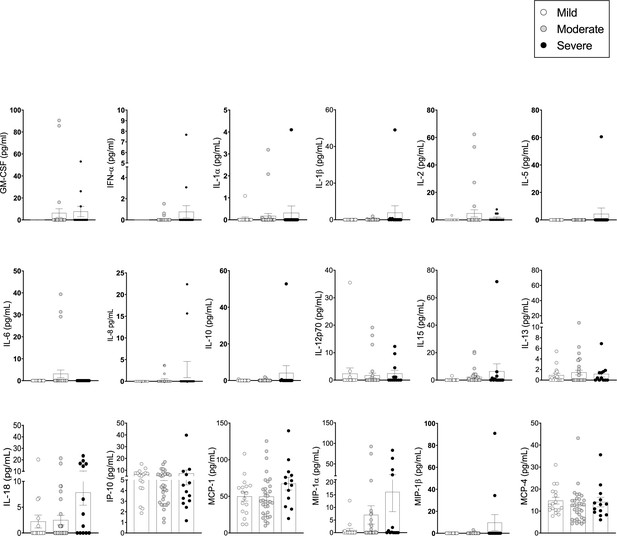
Pro-inflammatory cytokines/chemokines in the plasma of coronavirus disease-19 (COVID-19) patients at 3 months post admission.
Cytokines/chemokines were measured in the plasma of patients by Luminex. (A) Analyte levels (pg/ml) are shown for mild, moderate, and severe patients at 3 months post admission (63 samples: mild: N=17; moderate: N=32; severe: N=14, depicted in white, grey, and black symbols, respectively). Data are visualised as mean ± SEM. No statistical differences were detected for the cytokines included here, as calculated by one-way ANOVA test (Kruskal-Wallis test) with Dunn’s correction for multiple testing.
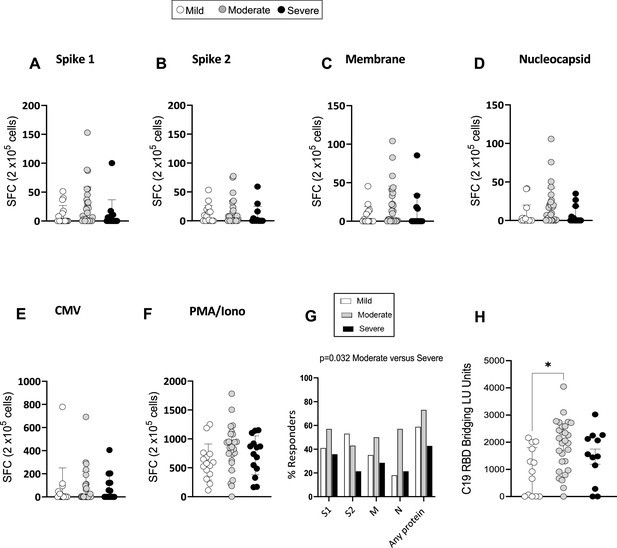
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific memory T-cell and antibody response at 3 months.
(A–F) Interferon gamma (IFN-γ) release measured by Enzyme-Linked Immune absorbent Spot (ELISpot) in peripheral blood mononuclear cells (PBMCs) from mild, moderate, and severe patients (N=61) upon stimulation with 15-mer peptide pools spanning SARS-CoV-2 spike 1 (A), spike 2 (B), membrane (C), nucleocapsid (D), cytomegalovirus (CMV) pp65 (E), and PMA/ionomycin (F). Results are shown as spot forming cells (SFC) relative to 2×105 PBMCs. (G) Percentages of responders assessed as patients from each severity group who displayed a response to the indicated peptide pool >5 SFC/2×105 PBMCs. (H) SARS-CoV-2 receptor binding domain (RBD) antibody titers in patients expressed as RBD bridging LU units. Data in A–F are visualised as mean ± SEM. Statistics were calculated by one-way ANOVA (Kruskal-Wallis test) with Dunn’s correction for multiple testing.
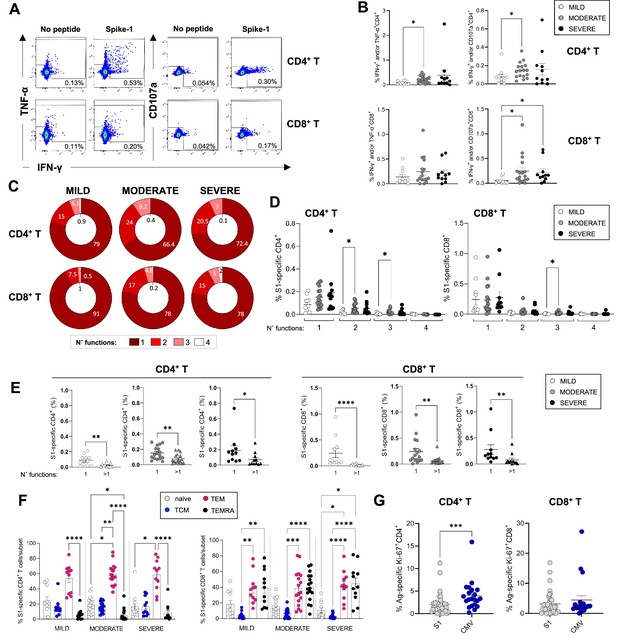
Magnitude and cytokine profiles of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific CD4+ and CD8+ T-cells at 3 months.
CD4+ and CD8+ T-cell responses targeting spike peptides were assessed by intracellular cytokine staining (ICS) in mild (N=11), moderate (N=17), and severe (N=11) patients. (A, B) Shown are representative flow cytometry plots of interferon gamma (IFN-γ) and tumor necrosis factor-alpha (TNF-α) or CD107a production by CD4+ and CD8+ T-cells (A) and the percentages of CD4+ (top panel) and CD8+ T (bottom panel) cells producing IFN-γ and/or TNF-α and IFN-γ and/or CD107a in the presence of spike-1 peptides (B). (C) Pie charts summarising the multifunctionality of T-cells specific for spike-1, defined as their capacity to produce 1, 2, 3, or 4 cytokines/CD107a (no. functions). (D) Spike-1 (S1) specific CD4+ (left panel) and CD8+ T-cells (right panel) that express 1–4 functions in mild, moderate, and severe patients. (E) Monofunctionality and polyfunctionality (>1 function) of CD4+ and CD8+ T-cells targeting spike-1 peptides in mild, moderate, and severe patients. (F) Expression of differentiation markers CD45RA/CCR7 by spike-1specific CD4+ and CD8+ T-cells in mild, moderate, and severe patients. Naïve cells = CCR7+CD45RA+ (white); T central memory cells (TCM)=CCR7+ CD45RA- (blue); T effector memory cells (TEM)=CCR7+ CD45RA- (red); T effector memory RA re-expressing cells (TEMRA)=CCR7+ CD45RA- (black). Data not significantly different between patient groups. (G) Percentage of spike-1-specific or CMV-specific CD4+ (left panel) and CD8+ T (right panel) cells that express Ki67. Data in A–B, D–G are visualised as mean ± SEM. Statistics were calculated by one-way ANOVA (Kruskal-Wallis test) with Dunn’s correction for multiple testing or by Mann-Whitney t-test.
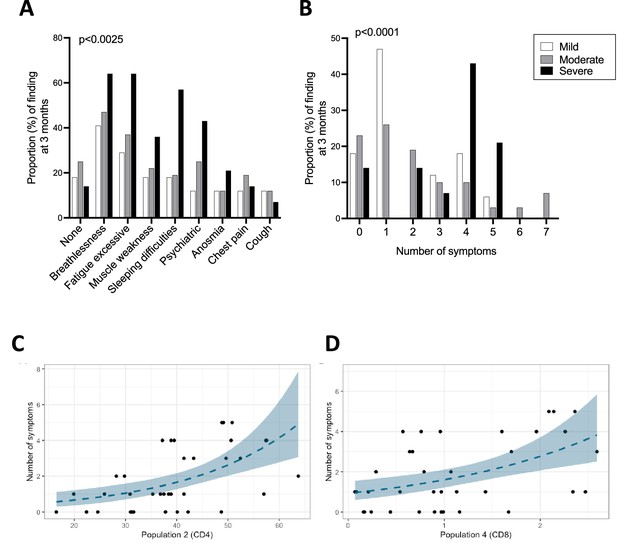
Ongoing symptoms at 3 months and associations with immune profiles.
(A, B) The percentage of patients with mild (N=17), moderate (N=32), and severe (N=14) coronavirus disease-19 (COVID-19) who reported the indicated symptom (A) or number of symptoms (B) at 3 months are indicated with white, grey, and black bars, respectively. Statistics were calculated using a Chi-square test. (C, D) Graphs depicting the association between number of symptoms and uniform manifold approximation and projection (UMAP) T-cells clusters in Poisson models, specifically CD4+ T-cell cluster 2 (C) and CD8+ T-cell cluster 4 (D).
-
Figure 6—source data 1
Associations between immune parameters and symptoms, physical component summary (PCS) or mental component summary (MCS) scores at 3 months in either unadjusted (A) or adjusted (B) Poisson regression models.
p-Values are shown with or without FDR correction (fdr_p or p values, respectively) for both adjusted and unadjusted models. Highlighted in grey are the two immune parameters that significantly correlated with symptoms after FDR correction. Only parameters with FDR uncorrected p-values <0.05 are included in the tables.
- https://cdn.elifesciences.org/articles/85009/elife-85009-fig6-data1-v2.docx
Tables
Details of the patients included in the immunological analysis of this study.
| Disease severity (n) | Mild | Moderate | Severe | |
|---|---|---|---|---|
| (n=17) | (n=32) | (n=14) | ||
| Age (median, SD) | 53±14.5 | 58±12.6 | 61.5±10 | |
| Sex, % (n) | Female | 35.3 (6) | 31.2 (10) | 50 (7) |
| Male | 64.7 (11) | 68.5 (22) | 50 (7) | |
| Ethnicity, % (n) | Caucasian | 83.3% (14) | 78.1% (25) | 87.7% (12) |
| Asian | 11.7% (2) | 12.5% (4) | 14.3% (2) | |
| Black | 5.9% (1) | 6.3% (2) | 0 (0) | |
| Missing data | 0 (0) | 3.1% (1) | 0 (0) | |
| BMI, kg/m2, % (average, SD) | Healthy | 7.6% (22.3±0.57) | 12.5% (23.75±0.5) | 14.3% (23±0) |
| Overweight | 35.3% (26.6±1.5) | 28.1% (27±1.39) | 28.6% (28.75±0.5) | |
| Obese | 35.3% (32.8±1.47) | 46.9% (32.7±2.46) | 28.6% (35±2.1) | |
| Extremely obese | 11.8% (61±28.2) | 9.38% (46.3±6.02) | 28.6% (46±7.34) | |
| Missing data | 0 | 3.1% | 0 | |
| Comorbidity % (n) | None | 47.1% (8) | 43.75% (14) | 14.3% (2) |
| Heart disease | 23.5% (4) | 15.6% (5) | 14.3% (2) | |
| T1DM | 0 (0) | 3.1% (1) | 7.14% (1) | |
| T2DM | 5.88% (1) | 12.5% (4) | 14.3% (2) | |
| Hypertension | 17.65 (3) | 18.75% (6) | 50% (7) | |
| Chronic lung disease | 5.88% (1) | 21.88% (7) | 50% (7) | |
| Kidney disease | 5.88 (1) | 9.37% (3) | 14.3% (2) | |
| Mental health | 0 (0) | 6.25% (2) | 14.3% (2) | |
| Cancer | 5.88% (1) | 3.1% (1) | 7.14 (1) | |
| Asthma | 5.88% (1) | 0 (0) | 14.3% (2) | |
| Total obesity | 47.1% (8) | 53.1% (17) | 57.1% (8) | |
| Other | 47.1% (8) | 28.1% (9) | 50% (7) | |
| Hospital stay (average days, SD) | 3.3±1.99 | 7.8±5.16 | 12±6.67 | |
| Ongoing symptoms at 3 months (n, %) | None | 3 (17.65%) | 8 (25%) | 2 (14.3%) |
| Dyspnoea | 7 (41.2%) | 15 (47%) | 9 (64.3%) | |
| Excessive fatigue | 5 (29.4%) | 12 (37.5%) | 9 (64.3%) | |
| Muscle weakness | 3 (17.65) | 7 (22%) | 5 (35.7%) | |
| Sleeping difficulties | 3 (17.65) | 6 (18.75%) | 8 (57%) | |
| Psychiatric | 2 (11.8%) | 8 (25%) | 6 (42.9%) | |
| Anosmia | 2 (11.8%) | 4 (12.5%) | 3 (21.4%) | |
| Chest pain | 2 (11.8%) | 6 (18.75%) | 2 (14.3%) | |
| Cough | 2 (11.8%) | 4 (12.5%) | 1 (7.14%) | |
| Other | 5 (29.4%) | 4 (12.5%) | 3 (21.4%) | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-human CD4 (RPA-T4) | Biolegend | Cat# 300535 | FC (0.625:50) |
| Antibody | Mouse monoclonal anti-human HLA-DR (L243) | Biolegend | Cat# 307640 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD38 (HIT2) | Biolegend | Cat# 303528 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human Ki-67 (Ki-67) | Biolegend | Cat# 350505 | FC (3:50) |
| Antibody | Mouse monoclonal anti-human CD16 (3G8) | Biolegend | Cat# 302007 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD8 (SK1) | Biolegend | Cat# 344713 | FC (3:50) |
| Antibody | Mouse monoclonal anti-human CD56 (NCAM16.2) | BD Biosciences | Cat# 564849 | FC (0.5:50) |
| Antibody | Mouse monoclonal anti-human CD3 (UCHT1) | BD Biosciences | Cat# 557943 | FC (1:50) |
| Antibody | Mouse monoclonal anti-human IFN-γ (B27) | BD Biosciences | Cat# 560371 | FC (3:50) |
| Antibody | Mouse monoclonal anti-human CD3 (UCHT1) | BD Biosciences | Cat# 560835 | FC (2:50) |
| Antibody | Mouse monoclonal anti-human CD107a (H4A3) | Biolegend | Cat# 328610 | FC (0.75:50) |
| Antibody | Mouse monoclonal anti-human TNF-α (MAb11) | Biolegend | Cat# 502946 | FC (3:50) |
| Antibody | Mouse monoclonal anti-human CD163 (GHI/61) | Biolegend | Cat# 333631 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD14 (M5E2) | Biolegend | Cat# 301833 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD68 (Y1/82A) | Biolegend | Cat# 333811 | FC (5:50) |
| Antibody | Mouse monoclonal anti-human CD66b (G10F5) | Biolegend | Cat# 305122 | FC (5:50) |
| Antibody | Mouse monoclonal anti-human TCR γ/δ (B1) | Biolegend | Cat# 331209 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD16 (3G8) | Biolegend | Cat# 302017 | FC (0.5:50) |
| Antibody | Mouse monoclonal anti-human CD80 (2D10) | Biolegend | Cat# 305219 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human CD69 (FN50) | Biolegend | Cat# 310931 | FC (5:50) |
| Antibody | Mouse monoclonal anti-human CXCR3 (G025H7) | Biolegend | Cat# 353714 | FC (1:50) |
| Antibody | Mouse monoclonal anti-human CD45RA (HI100) | BD Biosciences | Cat# 561882 | FC (10:50) |
| Antibody | Mouse monoclonal anti-human CCR7 (G043H7) | Biolegend | Cat# 353226 | FC (2.5:50) |
| Antibody | Mouse anti-human CD19 (HIB19) | Biolegend | Cat# 302216 | FC (1:50) |
| Antibody | Rat anti-human IL-2 (MQ1-17H12) | Biolegend | Cat# 500322 | FC (2:50) |
| Antibody | Mouse anti-human CD8a (RPA-T8) | Biolegend | Cat# 301014 | FC (0.3:50) |
| Antibody | Mouse anti-human IL-15Rα (JM7A4) | Biolegend | Cat# 330207 | FC (2.5:50) |
| Antibody | Mouse monoclonal anti-human PD1 (EH12.1) | BD Biosciences | Cat# 612791 | FC (3:50) |
| Antibody | Mouse recombinant anti-human Granzyme B (QA16A02) | Biolegend | Cat# 372219 | FC (2.5:50) |
| Biological sample (human) | Peripheral blood mononuclear cells (PBMCs) | DISCOVER study, Bristol, UK | Frozen - isolated PBMCs | |
| Biological sample (human) | Plasma | DISCOVER study, Bristol, UK | Frozen plasma | |
| Peptide, recombinant protein | SARS-CoV-2 spike protein overlapping peptide library (custom made) | Mimotopes | N/A | |
| Peptide, recombinant protein | SARS-CoV-2 membrane protein overlapping peptide library (custom made) | Mimotopes | N/A | |
| Peptide, recombinant protein | SARS-CoV-2 nucleocapsid protein overlapping peptide library (custom made) | Mimotopes | N/A | |
| Peptide, recombinant protein | CMV pp65 protein (AD169 strain) overlapping peptide library (custom made) | Mimotopes | N/A | |
| Other | Spike-RBD Antibody Bridging LIPS assay | DOI: 10.3389/fimmu.2022.968317 | N/A | |
| Peptide, recombinant protein | Nano-Glo | Promega | Cat# N1150 | |
| Commercial assay or kit | Human IFN-γ ELISpot BASIC kit | Mabtech | Cat# 34202A | |
| Commercial assay or kit | ProcartaPlex Mix&\Match 23-plex | Invitrogen | Cat# PPX-23-MXWCXFA | |
| Commercial assay or kit | Pan T Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-096-535 | |
| Commercial assay or kit | CellTrace Violet Cell Proliferation Kit, for flow cytometry | Thermo Fisher Scientific | Cat# C34557 | |
| Commercial assay or kit | eBioscience Foxp3/Transcription factor fixation/permeabilisation buffer | Invitrogen | Cat# 00-5523-00 | |
| Commercial assay or kit | Zombie Aqua Fixable Viability Kit | Biolegend | Cat# 423102 | FC (1:1000) |
| Commercial assay or kit | Zombie NIR Fixable Viability Kit | Biolegend | Cat# 423105 | FC (1:100) |
| Commercial assay or kit | Dynabeads Human T-Activator CD3/CD28 | Thermo Fisher Scientific | Cat# 11131D | |
| Commercial assay or kit | OneComp eBeads Compensation Beads | Thermo Fisher Scientific | Cat# 01-1111-42 | |
| Commercial assay or kit | Human TruStain FcX | Biolegend | Cat# 422302 | FC (2.5:50) |
| Commercial assay or kit | Human Anti-Cytomegalovirus IgG ELISA Kit (CMV) | Abcam | Cat# ab108724 | |
| Software | FlowJo | BD | v10.8.1 | |
| Software | R | R Foundation for Statistical Computing | v4.0.4 | |
| Software | GraphPad Prism | GraphPad Software | v9.4 | |
| Software, algorithm | xPONENT | Software for Luminex Instruments | The basic xPONENT software | |
| Software, algorithm | BioSpot Software Suite | ImmunoSpot S6 Ultra-V Analyzer |





