Ancestral protein reconstruction reveals evolutionary events governing variation in Dicer helicase function
Figures
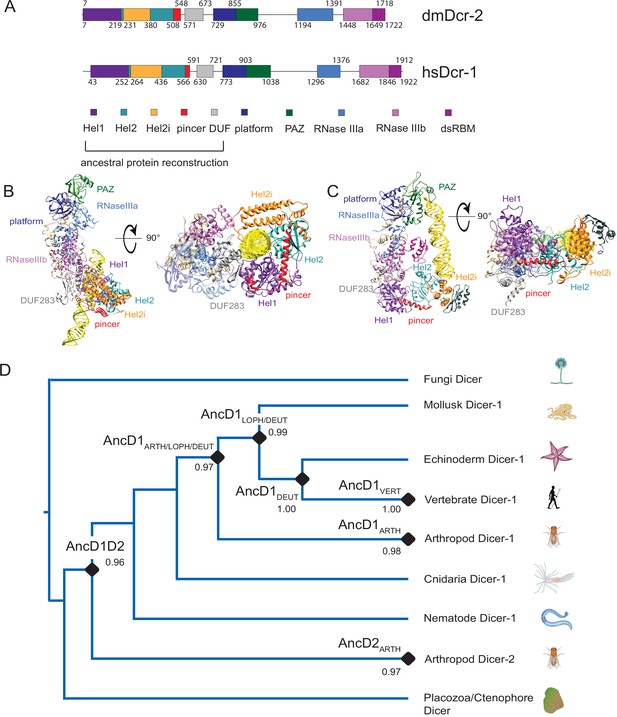
Phylogenetic analysis of Helicase domains and DUF283 of metazoan Dicer proteins.
(A) Domain organization of Drosophila melanogaster Dicer-2 (dmDcr2) and Homo sapiens Dicer (hsDcr), with colored rectangles showing conserved domain boundaries indicated by amino acid number. Domain boundaries were defined by information from NCBI Conserved Domains Database (CDD), available crystal and cryo-EM structures, and structure-based alignments from previous studies (Liu et al., 2018; Sinha et al., 2018; Marchler-Bauer et al., 2007). (B) Structure of dmDcr2 bound to dsRNA (yellow) at the helicase domain. Left: front view. Right: bottom-up view. (PDB: 7W0C). (C) Structure of hsDcr bound to dsRNA (yellow) at the platform/PAZ domain. Left: front view. Right: bottom-up view. (PDB: 5ZAL). (D) Summarized maximum likelihood phylogenetic tree constructed from metazoan Dicer helicase domains and DUF283. Nodes of interest are indicated with black rounded rhombi, and transfer bootstrap values are indicated. ARTH: arthropod, LOPH: lophotrochozoa, DEUT: deuterostome, and VERT: vertebrate.
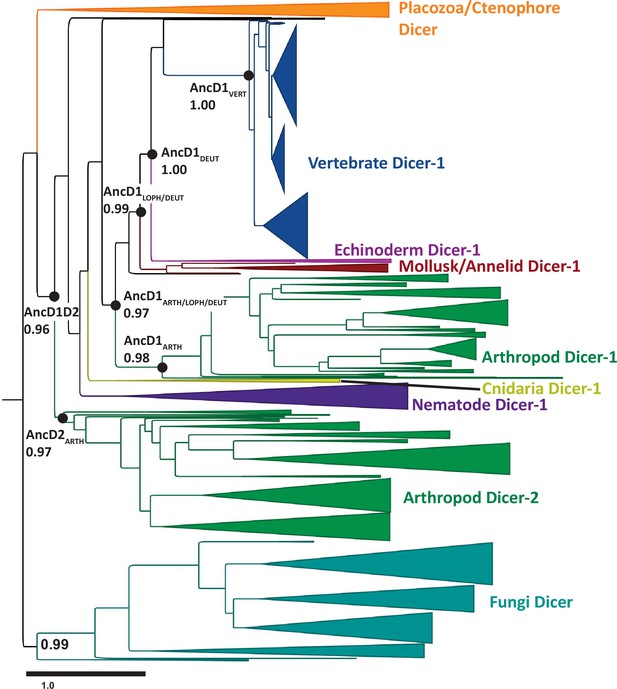
Maximum likelihood phylogenetic tree constructed from metazoan Dicer helicase domains and DUF283.
Dicer HEL-DUF phylogenetic tree visualized and annotated with FigTree. Resurrected ancestral nodes are indicated by black circles, with transfer bootstrap values indicated. Width of cartoon triangle base represents number of species. Scale bar represents total amino acid substitutions divided by the number of amino acid sites, i.e., amino acid substitutions per site.
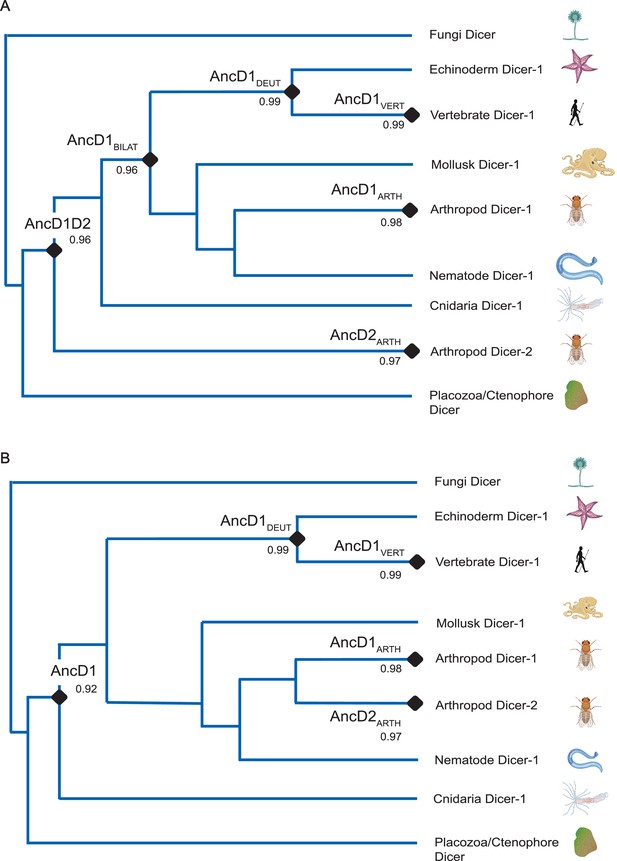
Alternative reconstructions of phylogenetic tree depicting Dicer HEL-DUF evolution.
(A) Summarized phylogenetic tree showing species-accurate relationships among bilaterian phyla. Gene duplication occurs early in animal evolution. Transfer bootstrap values at select nodes are indicated. BILAT, Bilateria. (B) Summarized phylogenetic tree showing species-accurate relationships among bilaterian phyla. Gene duplication is constrained to being arthropod specific. Transfer bootstrap values at select nodes are indicated.
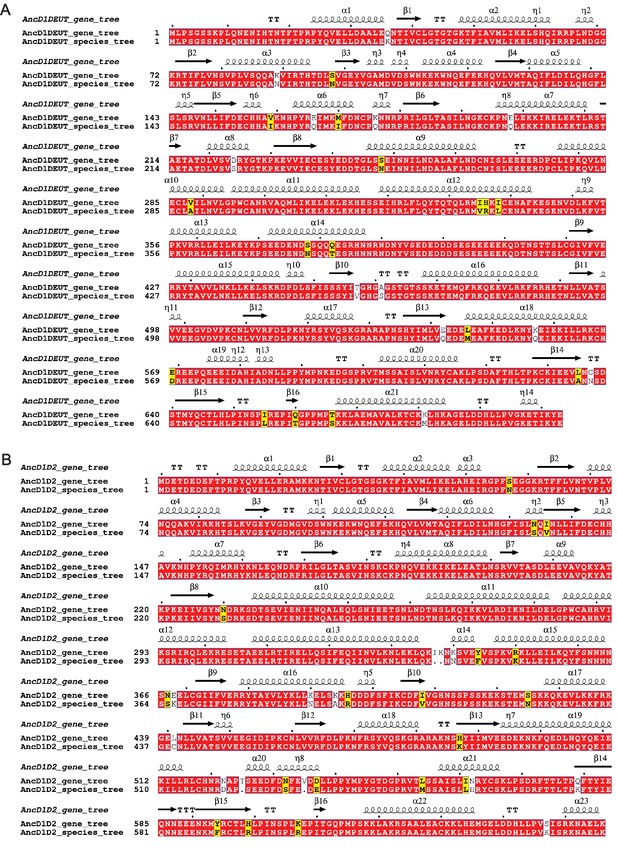
Constraining the phylogenetic tree to species-accurate relationships does not significantly impact ancestral protein reconstruction.
(A) Multiple sequence alignment illustrated with ESPript, depicting amino acid sequences for reconstructed AncD1DEUT node using either the gene tree or the species tree (Robert and Gouet, 2014). Red, identity; yellow, similarity; unshaded, no similarity. Secondary structures for AncD1DEUT determined with RosettaFold are shown above aligned sequences. TT represents beta turns. (B) Multiple sequence alignment illustrated with ESPript, depicting amino acid sequences for the reconstructed AncD1D2 node using either the gene tree or the species tree (Robert and Gouet, 2014). Red, identity; yellow, similarity; unshaded, no similarity. Secondary structures for AncD1D2 determined with RosettaFold are shown above aligned sequences. TT represents beta turns.
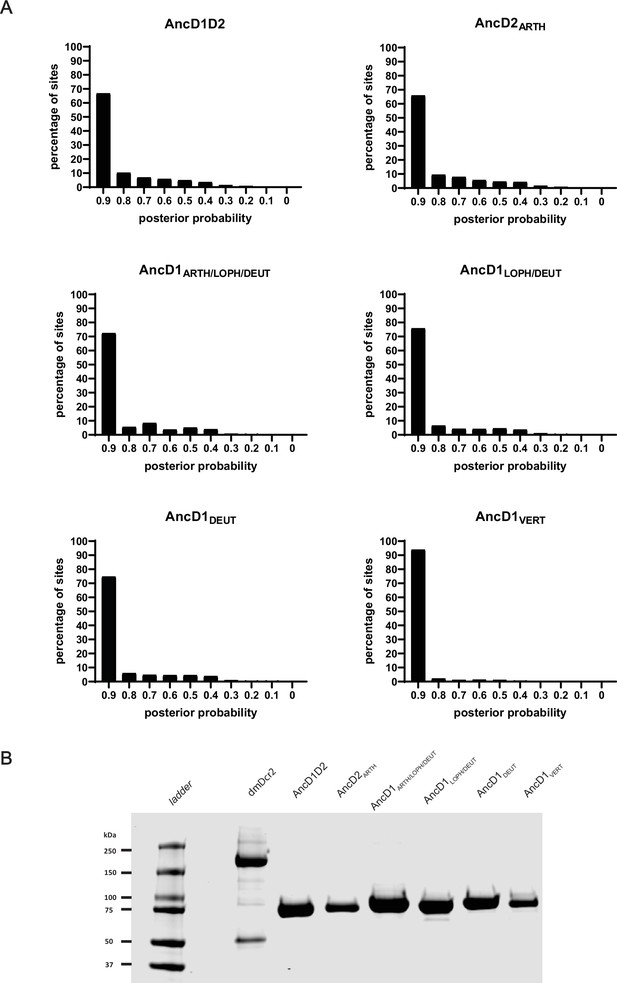
Reconstructed HEL-DUF constructs are predicted with high confidence and expressed recombinantly.
(A) Reconstructed HEL-DUFs at nodes of interest are predicted with posterior probabilities for each amino acid. Posterior probabilities for each amino acid are plotted and binned by 0.1. AncD1VERT is predicted with the highest confidence. (B) Coomassie-stained SDS-PAGE showing recombinantly expressed and purified full-length Drosophila melanogaster Dicer-2 and ancestral HEL-DUFs.
-
Figure 1—figure supplement 4—source data 1
Original digital image of SDS-PAGE gel used in B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig1-figsupp4-data1-v2.zip
-
Figure 1—figure supplement 4—source data 2
Posterior probabilities for ancestral states for all ancestrally reconstructed nodes in the maximum likelihood phylogeny.
Posterior probabilities represent each state in the ancestrally reconstructed protein prior to removal of low-probability insertions using an absence-presence alignment and the BIN model in RAXML-NG. Used in Figure 1—figure supplement 4.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig1-figsupp4-data2-v2.zip
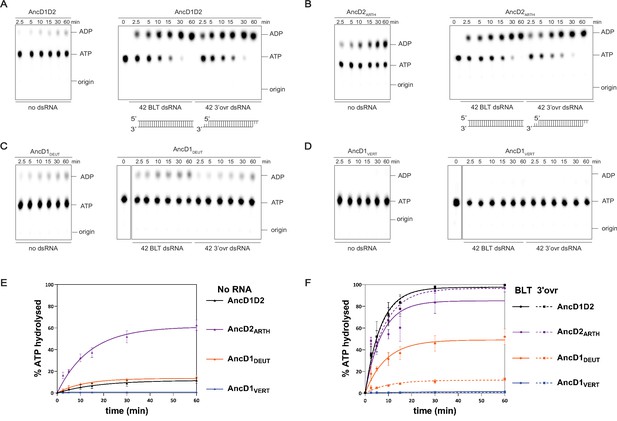
ATP hydrolysis capability is present in ancestral metazoan Dicer but lost at the common ancestor of vertebrates.
(A–D) PhosphorImages of representative thin-layer chromatography (TLC) plates showing hydrolysis of 100 µM ATP (spiked with α-32P-ATP) by 200 nM ancestral HEL-DUFs for various times as indicated, at 37°C, in the absence of dsRNA or in the presence of 400 nM 42 base-pair dsRNA with blunt (BLT) or 3’ovr termini (see cartoons; not radiolabeled). (E) Graph shows quantification of ATP hydrolysis assays (A–D) performed with select ancestral HEL-DUF enzymes in the absence of dsRNA. Data for ‘NO RNA’ reactions were fit to the pseudo-first order equation y=yo + A × (1-e−kt); where y=product formed (ADP in µM); A=amplitude of the rate curve, yo = baseline (~0), k=pseudo-first-order rate constant = kobs; t=time. Data points are mean ± SD (n≥3). (F) Graph shows quantification of ATP hydrolysis assays (A–D) performed with select ancestral HEL-DUF enzymes in the presence of dsRNA. Reactions with RNA were fit in two phases, first a linear phase for data below the first timepoint at 2.5 min, then a pseudo-first order exponential equation for remaining data. Equation, y=yo + A × (1-e−kt); where y=product formed (ADP in µM); A=amplitude of the rate curve, yo = baseline (~0), k=pseudo-first-order rate constant = kobs; t=time. Data points are mean ± SD (n≥3).
-
Figure 2—source data 1
Raw digital images of thin-layer chromatography plate used in 2A.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data1-v2.zip
-
Figure 2—source data 2
Raw digital image of thin-layer chromatography plate used in 2A.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data2-v2.zip
-
Figure 2—source data 3
Raw digital image of thin-layer chromatography plate used in 2B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data3-v2.zip
-
Figure 2—source data 4
Raw digital image of thin-layer chromatography plate used in 2B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data4-v2.zip
-
Figure 2—source data 5
Raw digital image of thin-layer chromatography plate used in 2C.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data5-v2.zip
-
Figure 2—source data 6
Raw digital image of thin-layer chromatography plate used in 2C.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data6-v2.zip
-
Figure 2—source data 7
Raw digital image of thin-layer chromatography plate used in 2D.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data7-v2.zip
-
Figure 2—source data 8
Raw digital image of thin-layer chromatography plate used in 2D.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig2-data8-v2.zip
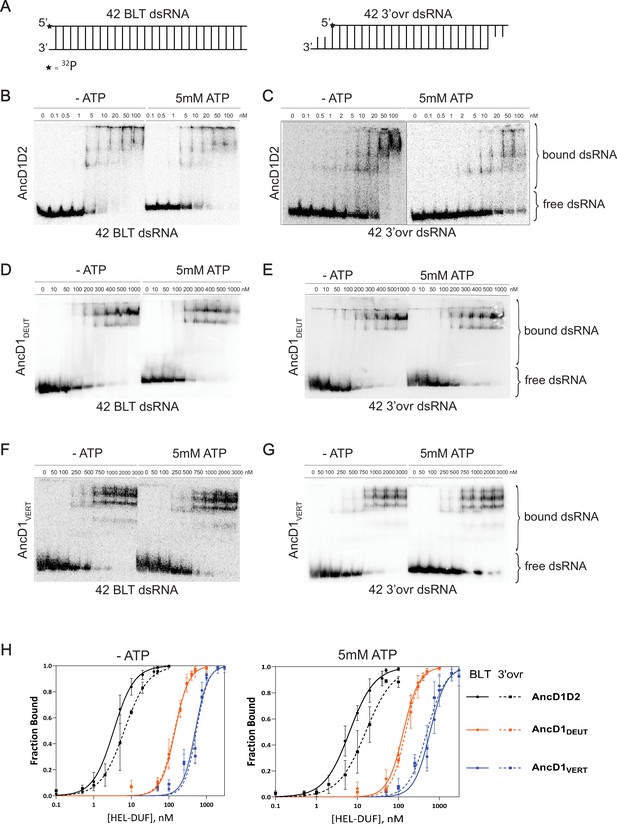
Binding affinity of ancestral HEL-DUF proteins to blunt (BLT) and 3’ovr dsRNA in the presence and absence of ATP.
(A) Cartoon of dsRNAs used in (B–G) showing position of 5’ 32P (*) on top, sense strand. (B–G) Representative PhosphorImages showing gel mobility shift assays using select ancestral HEL-DUF constructs, as indicated, and 42-base pair BLT or 3’ovr dsRNA in the absence (-) or presence of 5 mM ATP at 4ºC. (H) Radioactivity in PhosphorImages as in A–G was quantified to generate binding isotherms for ancestral HEL-DUF proteins. Fraction bound was determined using radioactivity for dsRNAfree and dsRNAbound. Data were fit to calculate dissociation constant, Kd, using the Hill formalism, where fraction bound = 1/(1 + [Kdn/[P]n]). Data points, mean ± SD (n≥3).
-
Figure 3—source data 1
Raw digital image of Gel Shift phosphoimager plate used in 3B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data1-v2.zip
-
Figure 3—source data 2
Raw digital image of Gel Shift phosphoimager plate used in 3C.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data2-v2.zip
-
Figure 3—source data 3
Raw digital image of Gel Shift phosphoimager plate used in 3C.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data3-v2.zip
-
Figure 3—source data 4
Raw digital image of Gel Shift phosphoimager plate used in 3D.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data4-v2.zip
-
Figure 3—source data 5
Raw digital image of Gel Shift phosphoimager plate used in 3E.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data5-v2.zip
-
Figure 3—source data 6
Raw digital image of Gel Shift phosphoimager plate used in 3F.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data6-v2.zip
-
Figure 3—source data 7
Raw digital image of Gel Shift phosphoimager plate used in 3G.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig3-data7-v2.zip
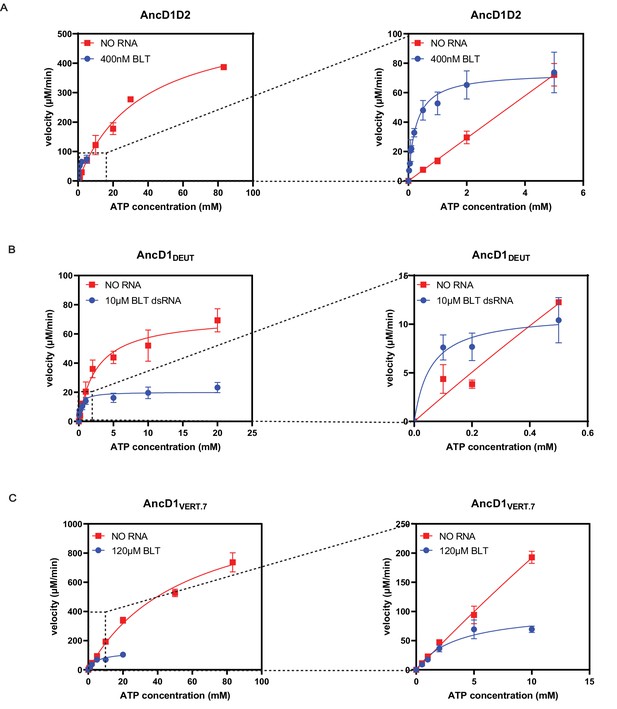
Blunt (BLT) dsRNA improves efficiency of ATP hydrolysis by improving affinity of ATP to ancient HEL-DUF enzymes.
(A) Michaelis-Menten plots for basal and dsRNA-stimulated ATP hydrolysis by AncD1D2. Basal ATP hydrolysis measured at 500 nM AncD1D2, while dsRNA-stimulated hydrolysis is measured at 100 nM. Velocities for dsRNA-stimulated reaction have been multiplied by five to normalize this concentration difference. Right: inset showing Michaelis-Menten plot at low ATP concentrations. Hydrolysis data for individual ATP concentrations is included in Figure 4—figure supplement 1. (B) Michaelis-Menten plots for basal and dsRNA-stimulated ATP hydrolysis by 500 nM AncD1DEUT. Right: inset showing Michaelis-Menten plot at low ATP concentrations. Hydrolysis data for individual ATP concentrations is included in Figure 4—figure supplement 1. (C) Michaelis-Menten plots for basal and dsRNA-stimulated ATP hydrolysis by 5 µM AncD1VERT.7. Right: inset showing Michaelis-Menten plot at low ATP concentrations. Hydrolysis data for individual ATP concentrations is included in Figure 4—figure supplement 1. Data points, mean ± SD (n≥3).
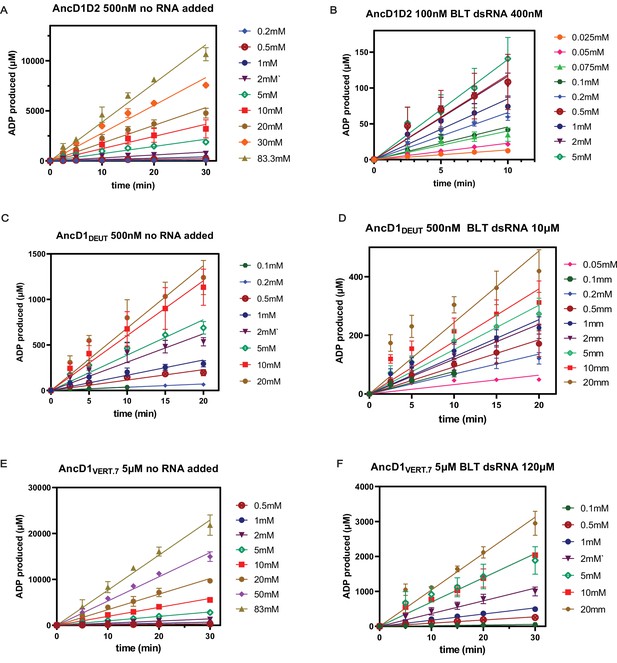
Plots of ADP production over time for ancestral HEL-DUF constructs.
(A) Basal ATP hydrolysis by 500 nM AncD1D2, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. (B) dsRNA-stimulated ATP hydrolysis by 100 nM AncD1D2 and 400 nM blunt (BLT) dsRNA, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. (C) Basal ATP hydrolysis by 500 nM AncD1DEUT, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. (D) dsRNA-stimulated ATP hydrolysis by 500 nM AncD1DEUT and 10 µM BLT dsRNA, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. (E) Basal ATP hydrolysis by 5 µM AncD1VERT.7, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. (F) dsRNA-stimulated ATP hydrolysis by 5 µM AncD1VERT.7 and 120 µM BLT dsRNA, measured by linear ADP production over time for indicated ATP concentrations. Velocity of each reaction is the slope of the line. Data points, mean ± SD (n≥3).
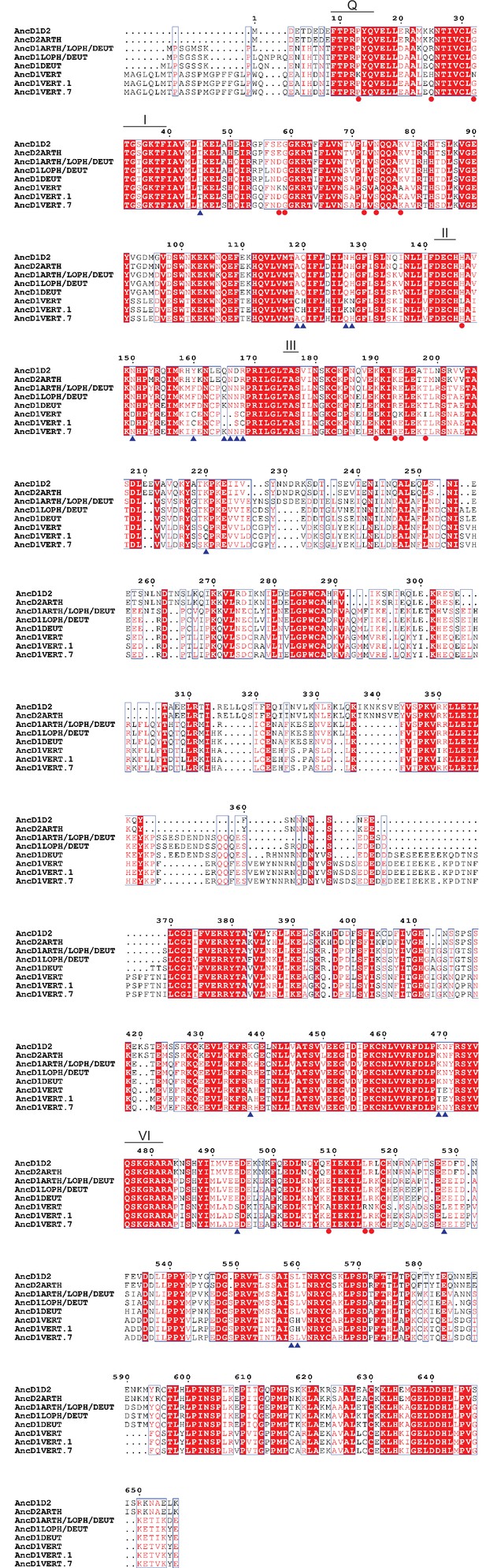
Multiple sequence alignment of ancestral HEL-DUF constructs and AncD1VERT rescue constructs.
Multiple sequence alignment for ancestral HEL-DUF constructs and vertebrate HEL-DUF rescue constructs, carried out with PRANK and illustrated with ESPript. Red shading/white text indicates identity, no shading/red text indicates similarity, and black text indicates no conservation. Columns with black and red text have at least 70% conservation, represented by red text, while black text indicates the non-conserved or variant amino acids. Amino acid substitutions in both rescue constructs are indicated by red circles below the column, while amino acid changes specific to AncD1VERT.7 are indicated by blue triangles. Residues numbered using shortest ancestral HEL-DUF amino acid sequence. Motif Q is numbered 9–16, motif I numbered 31–38, motif II numbered 142–145, motif III numbered 175–177, and motif VI numbered 476–482.
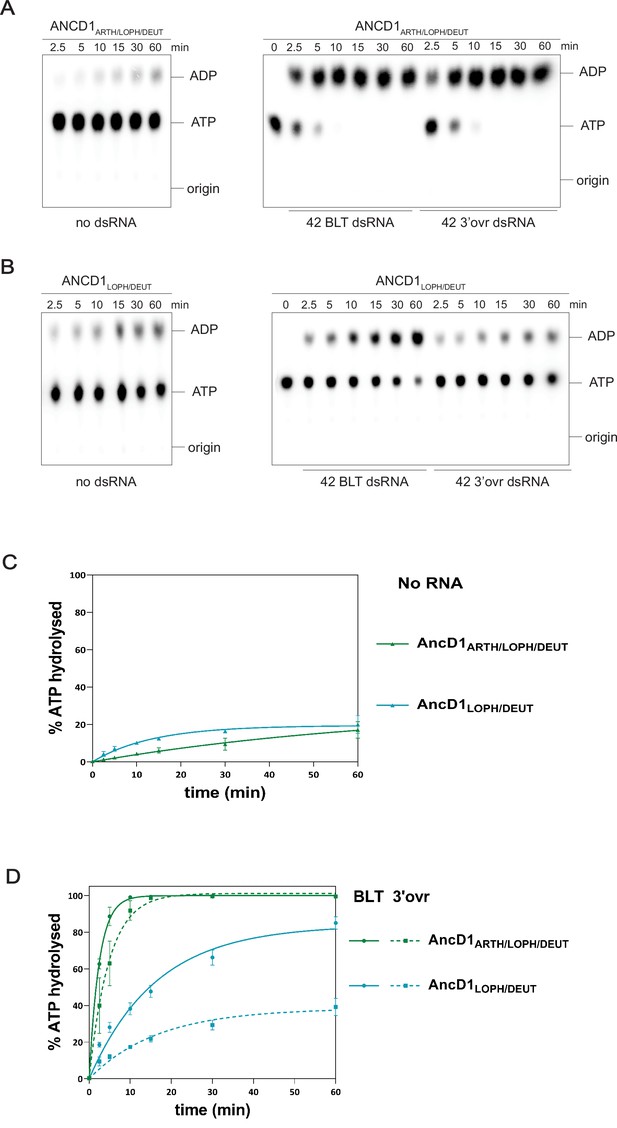
ATP hydrolysis of ancestral HEL-DUF proteins reconstructed from incongruent nodes.
(A–B) PhosphorImages of representative thin-layer chromatography (TLC) plates showing hydrolysis of 100 µM ATP (spiked with α-32P-ATP) by 200 nM AncD1ARTH/LOPH/DEUT (A) or AncD1LOPH/DEUT (B) in the absence of dsRNA (left) or in the presence of 400 nM 42 base-pair dsRNA with blunt (BLT) or 3’ 2-nucleotide overhang (right). (C–D) Graph shows quantification of ATP hydrolysis assays in A–B performed with select ancestral HEL-DUF enzymes in the absence (C) or presence (D) of dsRNA. Data for ‘NO RNA’ reactions were fit to the pseudo-first order equation y=yo + A × (1-e−kt); where y=product formed (ADP in µM); A=amplitude of the rate curve, yo = baseline (~0), k=pseudo-first-order rate constant = kobs; t=time. Reactions with RNA were fit in two phases, first a linear phase for data below the first timepoint at 2.5 min, then a pseudo-first order exponential equation for remaining data. Data points are mean ± SD (n≥3).
-
Figure 4—figure supplement 3—source data 1
Raw digital image of thin-layer chromatography plate used in Figure 4—figure supplement 3A, left panel.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp3-data1-v2.zip
-
Figure 4—figure supplement 3—source data 2
Raw digital image of thin-layer chromatography plate used in Figure 4—figure supplement 3A, right panel.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp3-data2-v2.zip
-
Figure 4—figure supplement 3—source data 3
Raw digital image of thin-layer chromatography plate used in Figure 4—figure supplement 3B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp3-data3-v2.zip
-
Figure 4—figure supplement 3—source data 4
Raw digital image of thin-layer chromatography plate used in Figure 4—figure supplement 3B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp3-data4-v2.zip
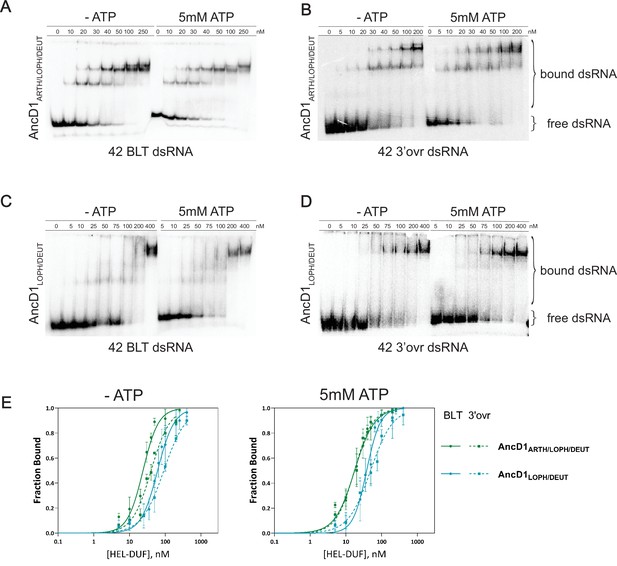
Affinity of AncD1ARTH/LOPH/DEUT and AncD1LOPH/DEUT for binding blunt (BLT) and 3’ovr dsRNA in the absence and presence of ATP.
(A–D) Representative PhosphorImages showing gel mobility shift assays using select ancestral HEL-DUF constructs as indicated, and 42 base-pair BLT or 3’ovr dsRNA in the absence or presence of 5 mM ATP. (E) Radioactivity in PhosphorImages as in A–D was quantified to generate binding isotherms for ancestral HEL-DUF proteins. Fraction bound was determined using radioactivity for dsRNAfree and dsRNAbound. Data were fit to calculate dissociation constant, Kd, using the Hill formalism, where fraction bound = 1/(1 + [Kdn/[P]n]). Data points, mean ± SD (n≥3).
-
Figure 4—figure supplement 4—source data 1
Raw digital image of Gel Shift phosphoimager plate used in Figure 4—figure supplement 4A.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp4-data1-v2.zip
-
Figure 4—figure supplement 4—source data 2
Raw digital image of Gel Shift phosphoimager plate used in Figure 4—figure supplement 4B.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp4-data2-v2.zip
-
Figure 4—figure supplement 4—source data 3
Raw digital image of Gel Shift phosphoimager plate used in Figure 4—figure supplement 4C.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp4-data3-v2.zip
-
Figure 4—figure supplement 4—source data 4
Raw digital image of Gel Shift phosphoimager plate used in Figure 4—figure supplement 4D.
- https://cdn.elifesciences.org/articles/85120/elife-85120-fig4-figsupp4-data4-v2.zip
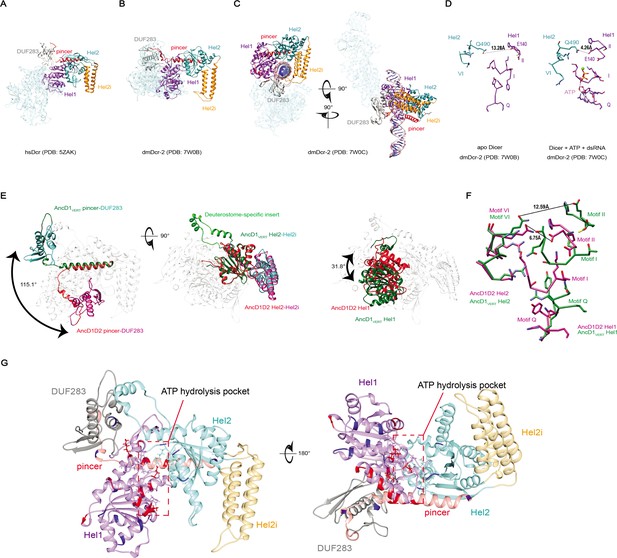
dsRNA binding triggers conformational changes in the HEL-DUF domains of Dicer.
(A) Bottom-up view of the structure of Homo sapiens Dicer (hsDcr) in the apo state (PDB: 5ZAK). Helicase subdomains and DUF283 are colored. Rest of enzyme is transparent. (B) Bottom-up view of the structure of dmDcr2 in the apo state (PDB: 7W0B). Helicase subdomains and DUF283 are colored for visibility. Rest of enzyme is transparent. (C) Structure of dmDcr2 bound to dsRNA in the ‘early translocation’ state (PDB: 7W0C). Helicase subdomains and DUF283 are colored for visibility. (D) Details of interactions at the ATP binding pocket of dmDcr2, comparing the distance between Motif II and Motif VI for the apo enzyme and the enzyme in the presence of ATP and dsRNA. Green sphere is magnesium ion, a cofactor in Sf2 helicase ATP hydrolysis. (E) Structural alignment of predicted structures for AncD1D2 and AncD1VERT HEL-DUFs showing conformational differences in position of Hel1 and pincer subdomains and DUF283. Pincer and DUF293, left panel; Hel2 and Hel2i, middle panel; Hel1, right panel. Green and teal coloring represent AncD1VERT subdomains, and red and violet coloring represent AncD1D2 subdomains. Deuterostome-specific insert refers to a Hel2 insertion present in AncD1DEUT and AncD1VERT. Structural predictions were performed with RosettaFold and AlphaFold2. pLDDT score: 81.76 for AncD1D2, 74.60 for AncD1VERT. (F) Details of the interactions of the ATP binding pocket for AncD1D2 and AncD1VERT, showing a wider cleft between Motif II and Motif VI for AncD1VERT (violet) compared to AncD1D2 (green). (G) RosettaFold predicted structures for AncD1VERT (transparent) showing sites of amino acid substitutions for both AncD1VERT.1 and AncD1VERT.7 marked in red, and amino acid substitutions unique to AncD1VERT.7 marked in blue. ATP hydrolysis pocket is depicted.
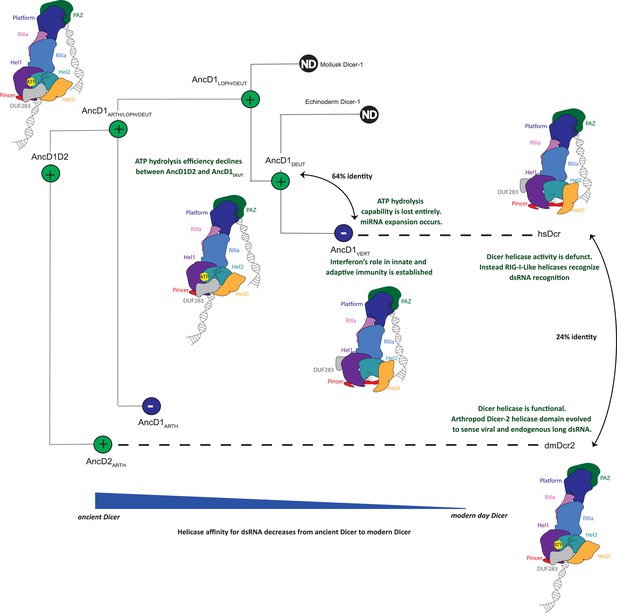
Model of metazoan Dicer evolution showing transition from a two-site dsRNA binding in ancestral Dicer to a 1-site dsRNA binding state in extant vertebrate and arthropod Dicers.
Early animals possessed one promiscuous Dicer enzyme capable of using both platform/PAZ and helicase domains for dsRNA recognition. After gene duplication, arthropod Dicer-2’s helicase domain becomes specialized for viral and endogenous long dsRNA processing and becomes the primary site of dsRNA binding. Deuterostome Dicer-1 may have retained two-site dsRNA recognition as helicase function declined, but at the onset of vertebrate evolution, Dicer-1 loses helicase function entirely and exclusively uses the platform/PAZ domain for dsRNA recognition.
Tables
Summary of kinetic data for ATP hydrolysis with 100 µM ATP.
| Construct | kburst (µM/min), NO RNA kobs (min–1) | kburst (µM/min), BLT dsRNA kobs (min–1) | kburst (µM/min), 3’ovr dsRNA kobs (min–1) |
|---|---|---|---|
| AncD1D2 | - 0.06±0.01 | 14.3±1.7 0.11±0.03 | 13.9±0.5 0.11±0.02 |
| AncD2ARTH | 6.47±0.8 0.05±0.01 | 19.3±0.9 0.04±0.02 | 14.6±2.3 0.08±0.02 |
| AncD1ARTH/LOPH/DEUT | - 0.01±0.01 | 25.1±0.7 0.41±0.02 | 16.0±3.8 0.21±0.04 |
| AncD1LOPH/DEUT | - 0.07±0.02 | 7.4±0.3 0.04±0.01 | 3.8±0.6 0.03±0.01 |
| AncD1DEUT | - 0.09±0.02 | 6.0±0.7 0.06±0.03 | 1.4±0.03 0.06±0.02 |
| AncD1VERT | - | - | - |
Dissociation constants for dsRNA binding to ancestral HEL-DUFs.
| Construct | Kd (nM) BLT, NO ATP Hill coefficient | Kd (nM) 3’ovr, NO ATP Hill coefficient | Kd (nM) BLT, 5 mM ATPHill coefficient | Kd (nM) 3’ovr, 5 mM ATP Hill coefficient |
|---|---|---|---|---|
| AncD1D2 | 3.4±0.4 1.6±0.2 | 6.5±0.8 1.4±0.2 | 6.4±0.7 1.4±0.2 | 15.9±2.4 1.3±0.2 |
| AncD2ARTH | n.d. | n.d. | n.d. | n.d. |
| AncD1ARTH/LOPH/DEUT | 23.8±2.2 2.0±0.4 | 40.1±3.7 1.7±0.3 | 17.5±2.1 1.6±0.3 | 17.3±1.5 1.7±0.2 |
| AncD1LOPH/DEUT | 60.9±5.9 1.9±0.3 | 90.8±8.8 1.4±0.2 | 38.0±3.5 2.3±0.5 | 49.0±5.2 1.4±0.2 |
| AncD1DEUT | 145.1±9.1 2.3±0.3 | 140.0±8.9 2.3±0.3 | 131.8±8.7 2.2±0.3 | 149.8±8.3 2.4±0.3 |
| AncD1VERT | 502.4±40.5 2.6±0.5 | 537.8±47.5 2.8±0.6 | 592±48.6 2.2±0.4 | 500.3±58.0 1.9±0.4 |
Michaelis-Menten parameters for steady state ATP hydrolysis reactions.
| Construct | kcat (min–1) | KM (µM) | kcat/KM (µM–1 min–1) |
|---|---|---|---|
| AncD1D2, no dsRNA | 1117±94.5 | 35812±6,367 | 0.031 |
| AncD1D2, BLT dsRNA | 147.8±6.3 | 256±67.5 | 0.577 |
| AncD1DEUT, no dsRNA | 144.1±14.9 | 2550±855 | 0.055 |
| AncD1DEUT, BLT dsRNA | 40.31±3.78 | 336.4±142 | 0.12 |
| AncD1VERT, no dsRNA | - | - | - |
| AncD1VERT.7, no dsRNA | 257.7±28.7 | 61739±12,886 | 0.004 |
| AncD1VERT.7, BLT dsRNA | 24.87±3.52 | 5173±1929 | 0.005 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pFastBac-OSF | Thermo Fisher Scientific | Cat# 10360014 | Modified in-house to add OSF tag |
| Cell line (Spodoptera frugiperda) | Sf9 | Expression Systems | Cat# 94–001 S | Suspension insect cells |
| Strain and strain background (Escherichia coli) | DH10Bac | Thermo Fisher Scientific | Cat #10361012 | Max Efficiency Competent Cells |
| Antibody | Anti-gp64-PE (mouse, monoclonal) | Expression Systems | Cat# 97–101 | Baculovirus Titering Kit |
| Chemical compound and drug | Cellfectin II | Thermo Fisher Scientific | Cat# 10362100 | Transfection reagent |
| Software and algorithm | RAXML-NG | RAXML-NG | RRID:SCR_022066 | |
| Sequence-based reagent | 42-nucleotide sense RNA | Integrated DNA Technologies (IDT) | Single-stranded RNA | GGGAAGCUCAGAAUA UUGCACAAGUAGAGC UUCUCGAUCCCC |
| Sequence-based reagent | 42-nucleotide BLUNT antisense RNA | IDT | Single-stranded RNA | GGGGAUCGAGAAGCU CUACUUGUGCAAUAU UCUGAGCUUCCC |
| Sequence-based reagent | 42-nucleotide 3’overhang antisense RNA | IDT | Single-stranded RNA | GGAUCGAGAAGCUCUA CUUGUGCAAUAUUCUG AGCUUCCCGG |
Additional files
-
Supplementary file 1
Fasta file containing amino acid sequences of ancestrally reconstructed proteins and engineered protein constructs.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp1-v2.txt
-
Supplementary file 2
Fasta file containing multiple sequence alignment file used as input for phylogeny construction and ancestral protein reconstruction.
Protein accession numbers from NCBI.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp2-v2.txt
-
Supplementary file 3
Text file containing the reconstructed ancestral states for all ancestral nodes in the maximum likelihood phylogenetic tree.
Reconstructions represent primary amino acid sequences prior to removal of low-probability insertions using binary states generated from the BIN model in RAXML-NG.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp3-v2.txt
-
Supplementary file 4
Text file containing the reconstructed ancestral states for all ancestral nodes in the maximum likelihood phylogenetic tree, using the multiple sequence alignment in the form of an absence-presence matrix and the BIN model in RAXML-NG for ancestral protein reconstruction.
One (1) represent the presence of amino acid residues, and zero (0) represents the absence. Protein accession numbers from NCBI.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp4-v2.txt
-
Supplementary file 5
Newick file of maximum likelihood metazoan Dicer HEL-DUF phylogeny used for ancestral reconstruction (shown in Figure 1 and Figure 1—figure supplement 1).
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp5-v2.txt
-
Supplementary file 6
Newick file of maximum likelihood phylogeny with bilaterian species constrained (shown in Figure 1—figure supplement 2A).
Protein accession numbers from NCBI.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp6-v2.txt
-
Supplementary file 7
Text files containing python scripts used to convert the amino acid multiple sequence alignment to binary alignment, overlay both alignments, and remove amino acids that correspond with absence (0) in the binary alignment.
- https://cdn.elifesciences.org/articles/85120/elife-85120-supp7-v2.txt
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85120/elife-85120-mdarchecklist1-v2.docx





