Reciprocal interactions between alveolar progenitor dysfunction and aging promote lung fibrosis
Figures
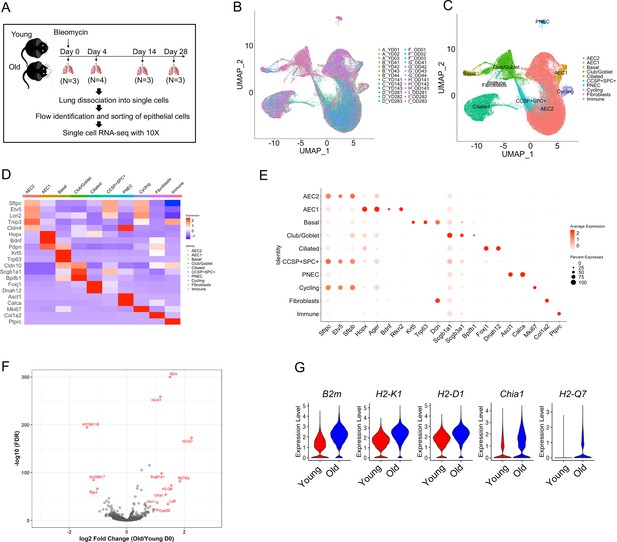
Transcriptome profiles of lung epithelial cells in young and old mice.
(A) Schematic of scRNA-seq analysis of flow sorted EpCAM+CD31−CD34−CD45− cells from lungs of uninjured (day 0) (n = 3), day 4 (n = 4), day 14 (n = 3), and day 28 post injury (n = 3) young and old mice. (B) Uniform Manifold Approximation and Projection (UMAP) visualization of 96,213 cells from all 26 samples. (C) UMAP visualization of epithelial cell clusters. (D) Heatmap of epithelial cell clusters. (E) Dot plots of conventional marker genes of epithelial cell clusters. (F, G) Gene expression in type 2 alveolar epithelial cells (AEC2s) from old vs young uninjured mice.
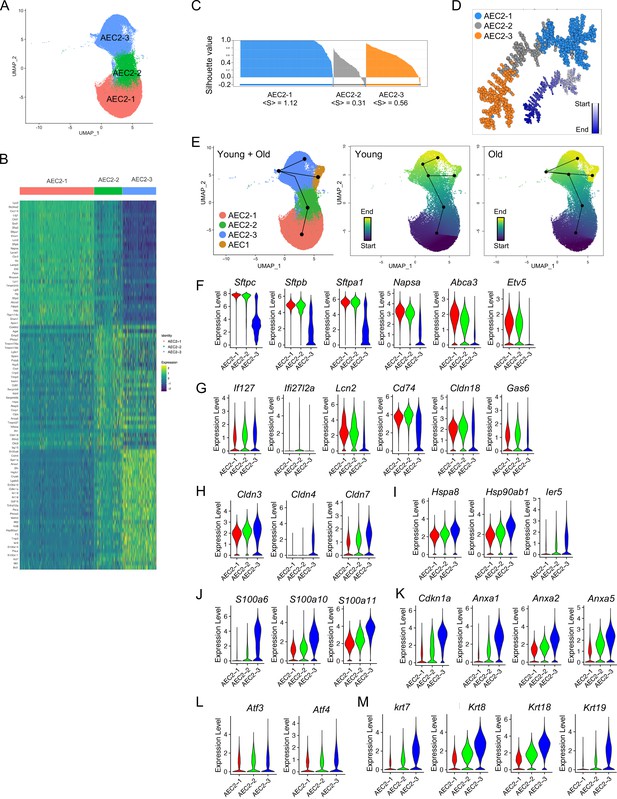
Definition of type 2 alveolar epithelial cell (AEC2) subsets.
(A) UMAP of 57,717 AEC2s showing in three subsets of AEC2s (red, AEC2-1; green, AEC2-2; and blue, AEC2-3). (B) Heatmap representing characteristics of three subsets of AEC2s. Each column represents the average expression value for one cell, grouped by cell cluster. Gene expression values are normalized in rows. (C) Correlation silhouette of AEC2 subsets. (D) Pseudotime analysis and correlation spanning tree of AEC2 subsets. (E) Slingshot trajectory inference analysis showed the lineage reconstructions of the AEC2 subsets and AEC1 clusters from young and old lungs. (F–M) Violin plots of gene expression in AEC2 subsets (red, AEC2-1; green, AEC2-2; and blue, AEC2-3). Expression of subset AEC2-1 marker genes (F), expression of subset AEC2-2 marker genes (G), expression of claudin family genes (H), expression of heat shock protein family genes (I), expression of S100 protein family genes (J), expression of Cdkn1a and annexin family genes (K), expression of Atf3 and Atf4 (L), and expression of the keratin family genes (M).
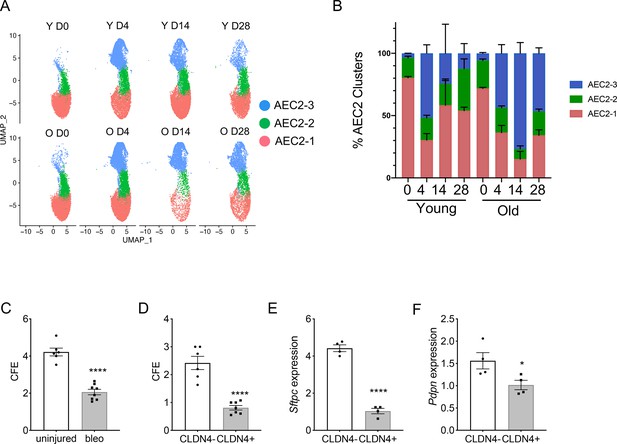
Type 2 alveolar epithelial cells (AEC2s) subsets were influenced by both aging and lung injury.
(A) UMAP showing the distributions of the three subsets of AEC2s grouped by age and injury date (red, AEC2-1; green, AEC2-2; and blue, AEC2-3. Y = young; O = old). (B) Percentage of AEC2 subsets grouped by age and injury date (red, AEC2-1; green, AEC2-2; and blue, AEC2-3). (C) Colony-forming efficiency (CFE) of AEC2s from uninjured and day 4 bleomycin-injured young mice (n = 6–8, ****p < 0.0001). (D) CFE of CLDN4+ and CLDN4− AEC2s isolated at day 14 after bleomycin treatment (n = 7–8, ****p < 0.0001). Sftpc (E) and Pdpn (F) expression of CLDN4+ and CLDN4− AEC2s derived from 3D cultured organoids by reverse transcription-polymerase chain reaction (RT-PCR) (n = 4, ****p < 0.0001, *p < 0.05). (C-F) Statistical anayses were by unpaired two-tailed Student’s t-test.
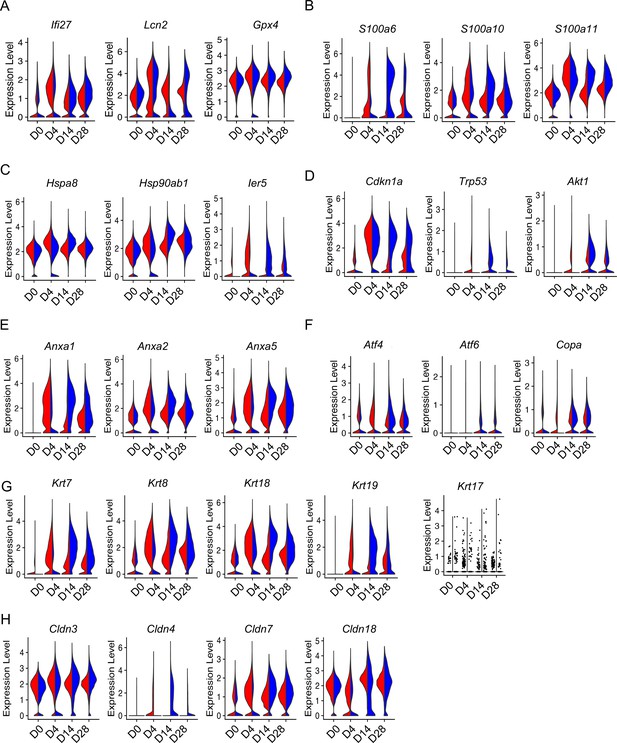
Aging enhanced type 2 alveolar epithelial cell (AEC2) injury with bleomycin time course.
Violin plots of gene expression in AEC2 from young and old mice at baseline day (D) 0 and days 4, 14, and 28 after bleomycin treatment. (A) Inflammatory and oxidative stress-related genes in AEC2s grouped by age and injury date; (B) S100a family genes; (C) heat shock protein family genes; (D) Senescence gene; (E) apoptosis-related genes; (F) ER stress-related genes; (G) keratin family genes; and (H) claudin family genes. Red, young; blue, old.
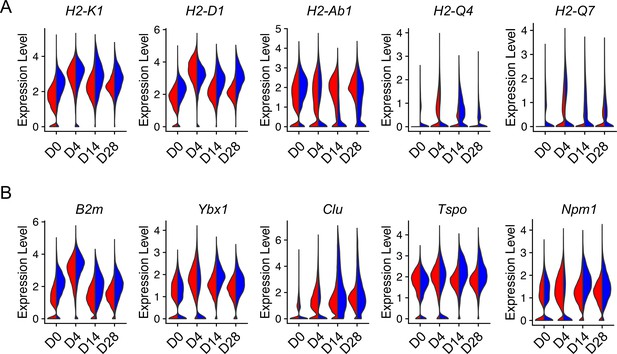
Injury-induced aging-related gene expression in type 2 alveolar epithelial cells (AEC2s).
Violin plots of aging-related gene expression in AEC2 from young and old mice at baseline day (D) 0 and days 4, 14, and 28 after bleomycin treatment. (A) Histocompatibility 2 genes. (B) Aging hallmark genes. Red, young; blue, old.
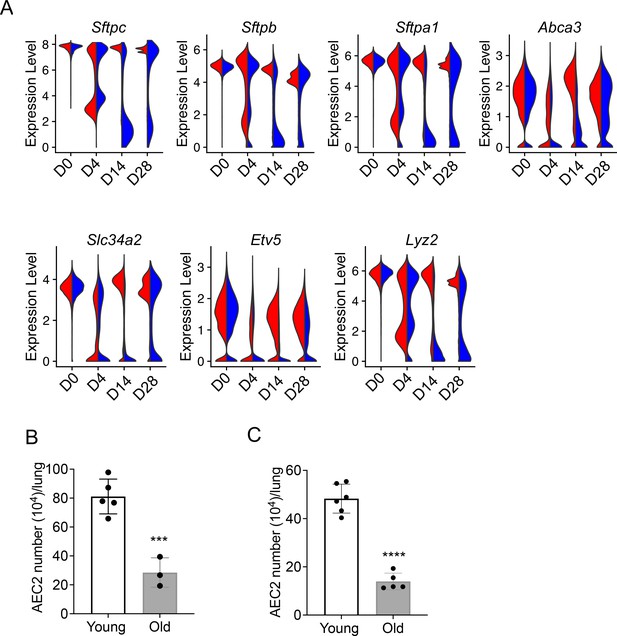
Decreased renewal capacity of injured aged type 2 alveolar epithelial cells (AEC2s).
(A) Violin plots of gene expression in AEC2 from young and old mice at baseline day (D) 0 and days 4, 14, and 28 after bleomycin treatment. Red, young; blue, old. (B) Number of AEC2s recovered from uninjured young and old mice (n = 5–6, ***p < 0.001, by unpaired two-tailed Student’s t-test). (C) Number of AEC2s recovered from bleomycin day 28 young and old mouse lungs (n = 3–5, ****p < 0.0001, by unpaired two-tailed Student’s t-test).
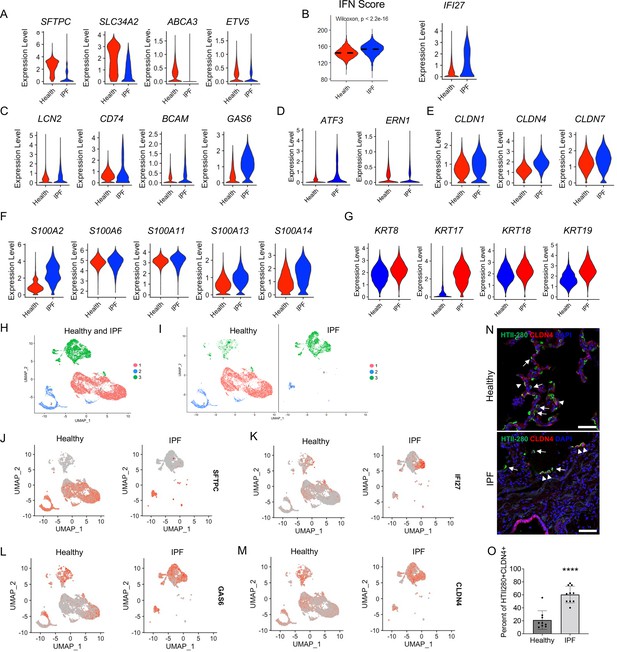
Idiopathic pulmonary fibrosis (IPF) type 2 alveolar epithelial cells (AEC2s) share the gene signatures of mouse AEC2 subsets AEC2-2 and AEC2-3.
(A) Violin plots of expression of mouse AEC2-1 signature genes in healthy and IPF AEC2s (red, healthy; blue, IPF AEC2). (B) IFN activation score and expression of IFI27 of AEC2s from healthy and IPF lungs (red, healthy; blue, IPF). (C) Violin plots of expression of mouse AEC2-2 signature genes in healthy and IPF AEC2s (red, healthy; blue, IPF AEC2). Violin plots of expression of AEC2-3 signature genes, including ER stress-related (D), claudin family (E), S100 family (F), and keratin family (G) in healthy and IPF AEC2s (red, healthy; blue, IPF). (H) UMAP showing three subsets of human AEC2s from healthy and IPF lungs. (I) AEC2 subset distribution in healthy and IPF AEC2s. Selected gene expression of AEC2 subsets from healthy and IPF lungs, SFTPC (J); IFI27 (K); GAS6 (L); and CLDN4 (M). (N) HTII-280 (arrows) and CLDN4 co-staining (arrowheads) of human lung sections, scale bars 50 μm. (O) Quantitation of the percentage of HTII280+CLDN4+ cells in total HTII-280+ AEC2s in healthy and IPF lungs (n = 9–10, ****p < 0.0001, by unpaired two-tailed Student’s t-test).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57Bl/6J | Jackson Laboratory | Strain #: 000664 RRID: IMSR_JAX:000664 | |
| Chemical compound, drug | Bleomycin | Hospira, Lake Forest, IL 60045 | NDC 61703-332-18 | 2.5 U/kg in vivo, mice |
| Antibody | Anti-Mouse EpCAM clone G8.8 (rat monoclonal) | BioLegend | Catalog # 118216 RRID: AB_1236471 | Flow 1:200 |
| Antibody | Anti-mouse CD24 clone M1/69 (rat monoclonal) | eBioscience | Catalog # 12-0242-82, RRID: AB_467169 | Flow 1:50 |
| Antibody | Anti-Sca-1 (Ly-6A/E)- clone D7 (rat monoclonal) | eBioscience | Catalog # 17-5981-82, RRID: AB_469487 | Flow 1:200 |
| Antibody | Anti-Mouse CD31 (PECAM-1) clone 390 (rat monoclonal) | eBioscience | Catalog # 13-0311-85, RRID: AB_466421 | Flow 1:40 |
| Antibody | Anti-Mouse CD34 clone RAM34 (rat monoclonal) | eBioscience | Catalog # 13-0341-85, RRID: AB_466425 | Flow 1:16 |
| Antibody | Anti-Mouse CD45 clone 30-F11 (rat monoclonal) | eBioscience | Catalog # 13-0451-85, RRID: AB_466447 | Flow 1:200 |
| Antibody | Anti-human CD31 clone WM59 (mouse monoclonal) | BioLegend | Clone WM59; RRID: AB_314327 | Flow 1:40 |
| Antibody | Anti-human CD45 clone WI30 (mouse monoclonal) | BioLegend | Catalog # 304016, RRID: AB_314404 | Flow 1:200 |
| Antibody | Anti-human EpCAM clone 9C4 (mouse monoclonal) | BioLegend | Catalog # 324212, RRID: AB_756086 | Flow 1:200 |
| Antibody | anti-human Claudin 4 IgG (rabbit polyclonal) | ProteinTech | 16195-1-AP, RRID: AB_2082969 | Flow 1:50; IF 1:200 |
| Antibody | anti-HT2-280 IgM (mouse monoclonal) | Terrace Biotech | TB-27AHT2-280, RRID: AB_2832931 | Flow 1:60; IF 1:200 |
| Sequence-based reagent | mouse Sftpc _F | This paper | PCR primers | GCAGGTCCCAGGAGCCAGTTC |
| Sequence-based reagent | mouse Sftpc_R | This paper | PCR primers | GGAGCTGGCTTATAGGCCGTCAG |
| Sequence-based reagent | mouse Pdpn_F | This paper | PCR primers | GCACCTCTGGTACCAACGCAGA |
| Sequence-based reagent | mouse Pdpn_R | This paper | PCR primers | TCTGAGGTTGCTGAGGTGGACAGT |
| Cell line (Mus musculus) | MLg2908, lung fibroblast (normal) | ATCC | Catalog CCL-206 | |
| Software, algorithm | Flow Jo | Tree Star | Version 9.9.6 | |
| Software, algorithm | RStudio | RStudio PBC | RRID: SCR_000432, version 2022.07.2 build 576 | |
| Software, algorithm | Prism | GraphPad | RRID: SCR_002798, version 8.4.3 |
Additional files
-
Supplementary file 1
Significantly enriched canonical pathways in AEC2 subsets using Ingenuity Pathway Analysis (IPA).
(a) Ingenuity pathway analysis showing activation Z-scores for selected pathways in AEC2-2 compared to AEC2-1. (b) Ingenuity pathway analysis showing activation Z-scores for selected pathways in AEC2-3 compared to AEC2-1.
- https://cdn.elifesciences.org/articles/85415/elife-85415-supp1-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85415/elife-85415-mdarchecklist1-v2.pdf





