Single cell preparations of Mycobacterium tuberculosis damage the mycobacterial envelope and disrupt macrophage interactions
Figures
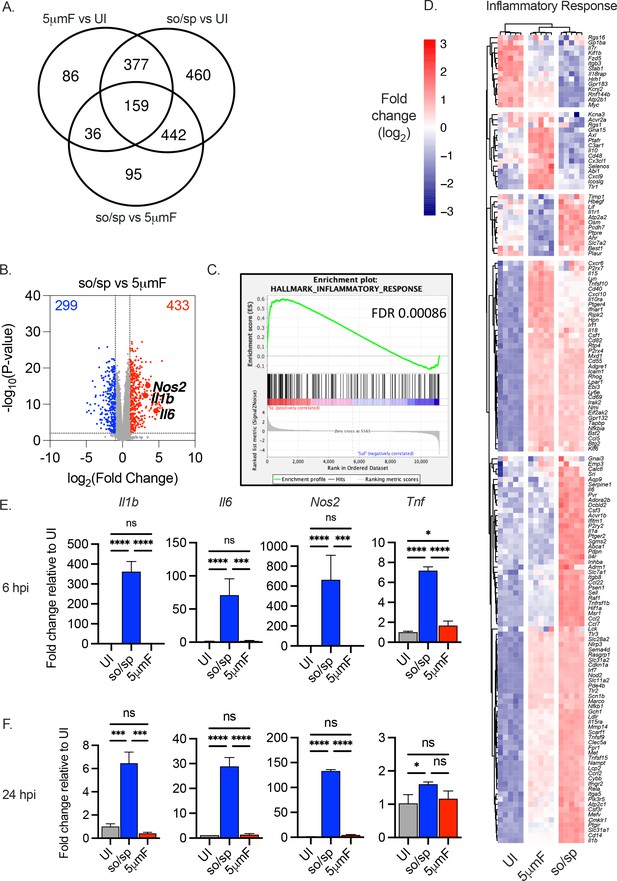
Cell preparation methods of Mtb impact macrophage responses.
(A–D) BMDMs were uninfected or infected with Mtb prepared by sonication and spin (so/sp) or filtration (5μmF) at an MOI of 5 and analyzed 72 hpi by RNA-seq (n=5 per condition). (A) Venn diagram illustrates the number of DEGs between samples. (B) Volcano plot shows genes differentially expressed in BMDMs infected with so/sp versus 5μmF Mtb. DEGs exhibiting an adjusted p-value of ≤0.01 and a linear fold change ≥2.00 (red) or ≤2.00 (blue) are indicated. (C–D) GSEA identified hallmark gene sets that were significantly enriched in so/sp- versus 5μmF-infected BMDMs (p≤0.01; FDR ≤0.01). Representative enrichment plot (C) and corresponding heat map (D) for the gene set ‘inflammatory response’. Expression values in heatmap were generated using log2 normalized CPM for each gene. (E and F) qPCR was performed on uninfected BMDMs or BMDMs infected with so/sp- or 5μmF-prepared Mtb 6 (E) and 24 (F) hpi using an MOI of 10. Data are shown as fold change in gene expression relative to uninfected BMDMs. Data shown are mean +/-SD from one representative experiment with three biological replicates per group and two technical replicates per sample. qPCR experiments were performed at least three independent times. Statistical significance was determined with one-way ANOVA using Tukey’s multiple comparisons test. (A–F) Error bars indicate mean +/-SD. ns not significant; *p<0.05; ***<0.001; ****<0.0001.
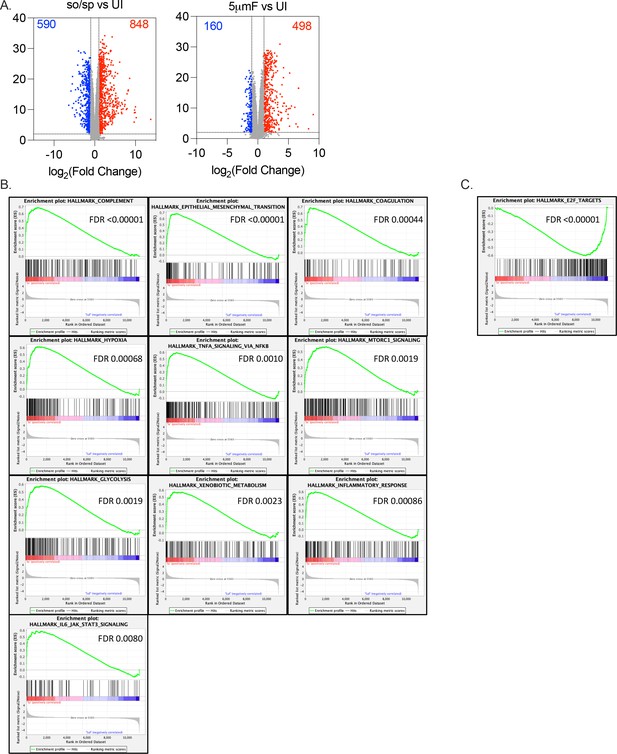
Infection-induced changes in macrophage gene expression depend on whether Mtb are sonicated or filtered.
(A) Volcano plot shows genes differentially expressed in BMDMs infected with so/sp versus UI (left) or 5μmF Mtb versus UI (right). DEGs exhibiting an adjusted p-value of ≤0.01 and a linear fold change ≥2.00 (red) or ≤–2.00 (blue) are indicated. (B–C) Gene set enrichment analysis (GSEA) leading edge graphs of hallmark gene sets that were enriched in BMDMs infected with so/sp relative to 5μmF Mtb (B) or 5μmF relative to so/sp (C).
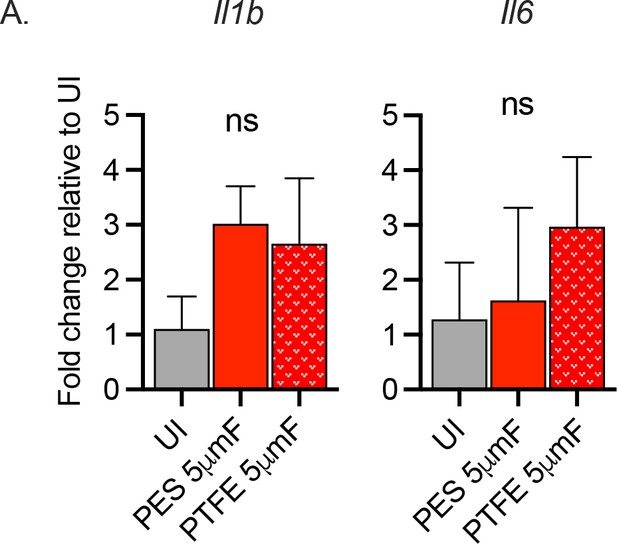
Filtered Mtb are uninflammatory irrespective of filter type.
BMDMs were uninfected or infected with Mtb prepared by passing through a 5μmF made of either PES or PTFE using a MOI of 10 and analyzed by qPCR 6 hpi. Data are presented as fold changes in gene expression relative to uninfected BMDMs. Data are representative of two to three experiments, each with three biological replicates per group and two technical replicates per sample. Statistical significance was determined with one-way ANOVA using Tukey’s multiple comparisons test. qPCR data are presented as fold change in gene expression relative to uninfected BMDMs. Error bars indicate mean +/-SD. ns not significant.
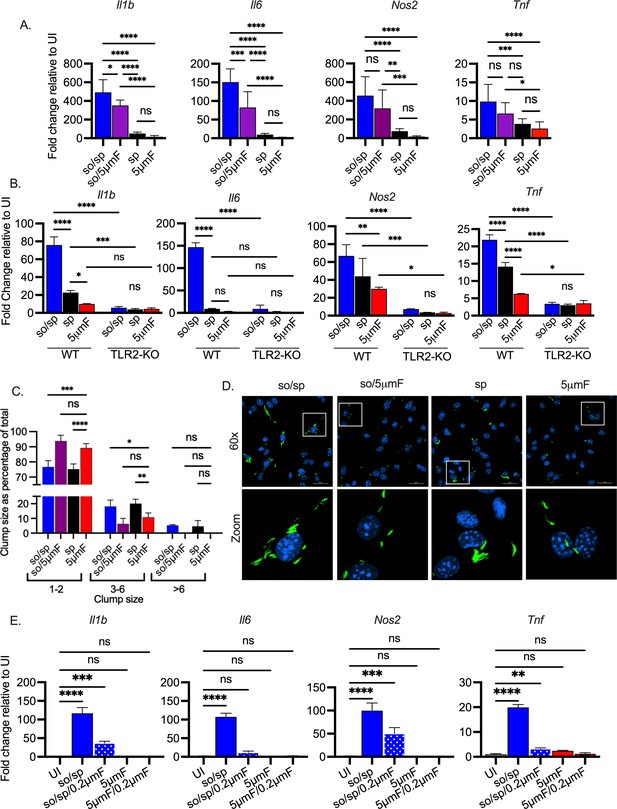
Sonicated bacteria induce high TLR2-dependent inflammatory responses.
(A) BMDMs were uninfected or infected with different preparations of Mtb as indicated at an MOI of 10 and analyzed by qPCR 6 hpi. Data are presented as fold changes in gene expression relative to uninfected BMDMs. Data are combined from two to three experiments, each with three biological replicates per group and two technical replicates per sample. Statistical significance was determined with one-way ANOVA using Tukey’s multiple comparisons test. (B) WT or Tlr2-/- BMDMs were uninfected or infected with different preparations of Mtb as indicated for 6 hr at an MOI of 10 and analyzed by qPCR. Data are presented as fold change in gene expression relative to uninfected BMDMs of the same mouse genotype. Data are representative of three experiments, each with three biological replicates per group and two technical replicates per sample. Statistical significance was determined with two-way ANOVA using Tukey’s multiple comparisons test. (C) BMDMs were infected with GFP-expressing bacteria (MOI 5) and visualized using immunofluorescence at 4 hpi. Bacteria were quantified and classified as single/doublets (1-2), small (3-6), or large (>6) clumps and quantified for each preparation. At least 100 bacterial occurrences of each preparation method were analyzed. Two-way ANOVA with Dunnett’s multiple comparisons test was used to assess statistical significance within each batch relative to the given 5µmF quantitation. (D) Representative fluorescence microscopy images of BMDMs (nuclei stained with DAPI) infected with GFP-expressing Mtb used in (C). Images are maximum-intensity projections. Boxed areas in the merged image are shown in higher magnification in the bottom panel. (E) BMDMs were untreated, infected with the indicated bacterial preparations at MOI 10, or treated with the sterile filtrate from different preparations for 6 hr and analyzed by qPCR. Data are presented as fold changes in gene expression relative to untreated BMDMs. Data are representative of 3 experiments, each with three biological replicates per group and two technical replicates per sample. Statistical significance was determined for each group relative to untreated BMDMs with one-way ANOVA using Dunnett’s multiple comparisons test. (A–B, D) Error bars indicate mean +/-SD. ns not significant; *p<0.05; **<0.01; ***<0.001; ****<0.0001.
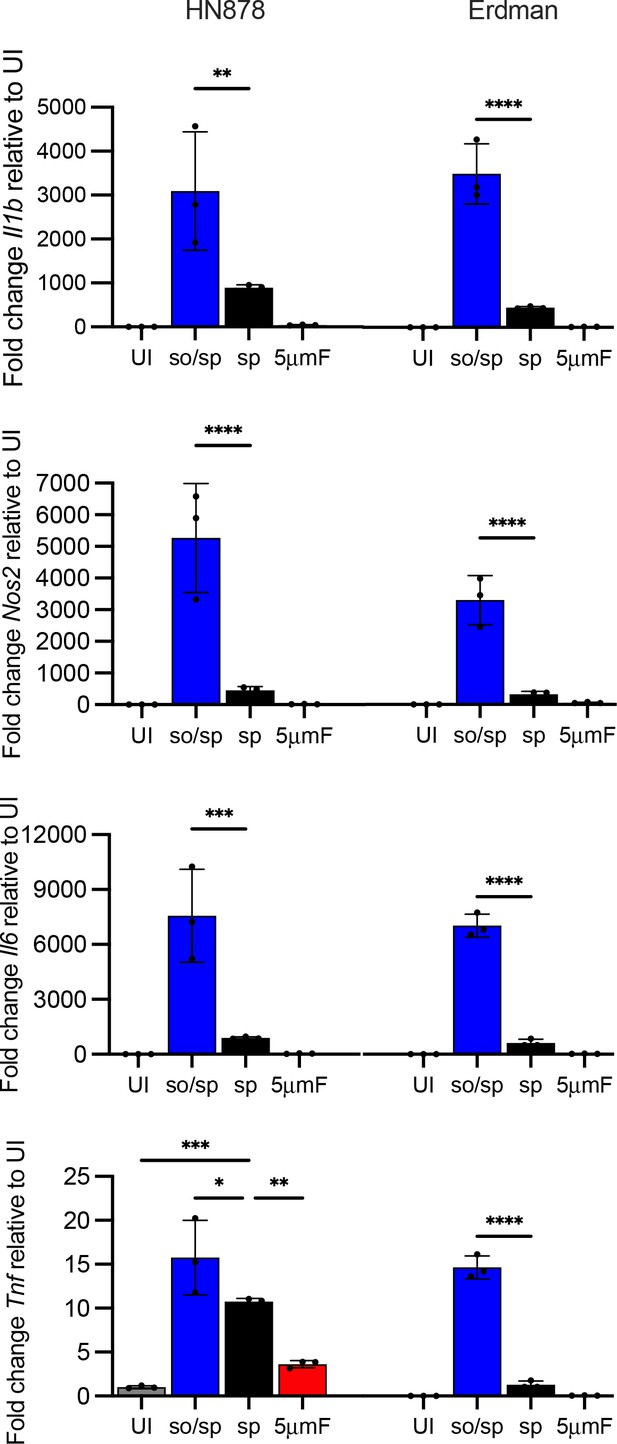
Sonicated HN878 and Erdman strains induce high TLR2-dependent inflammatory responses.
BMDMs were uninfected (UI) or infected with different preparations of HN878 or Erdman as indicated at an MOI of 10 and analyzed by qPCR 6 hpi. Data are presented as fold changes in gene expression relative to uninfected BMDMs. Three biological replicates were used per group and two technical replicates per sample. Statistical significance was determined with one-way ANOVA using Tukey’s multiple comparisons test. Error bars indicate mean +/-SD. ns not significant; *p<0.05; **p<0.01; ***<0.001; ****p<0.0001.
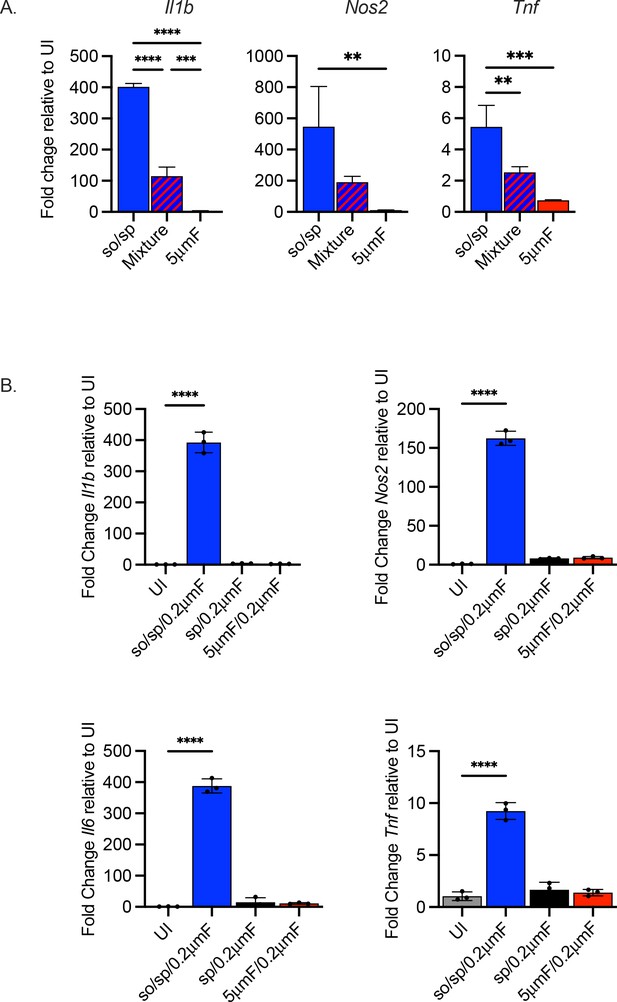
Filtered bacteria do not strongly inhibit response from sonicated bacteria and only the sterile filtrate of so/sp Mtb induces gene expression in BMDMs.
(A) BMDMs were uninfected or infected for 6 hr at a MOI of 10 with bacteria prepared by the so/sp or 5μmF-preparation or a mixture (1:1) of the two samples. Data are presented as fold change in gene expression relative to uninfected BMDMs. Statistical significance was determined with one-way ANOVA using Tukey’s multiple comparisons test. (B) BMDMs were untreated, or treated with the sterile filtrate from different preparations for 6 hr and analyzed by qPCR. Data are presented as fold changes in gene expression relative to untreated BMDMs. Statistical significance was determined for each group relative to untreated BMDMs with one-way ANOVA using Dunnett’s multiple comparisons test. (A–B) Data are representative of three experiments, each with three biological replicates per group and two technical replicates per sample. Error bars indicate mean +/-SD. ns not significant; **p<0.01; ***<0.001; ****<0.0001.
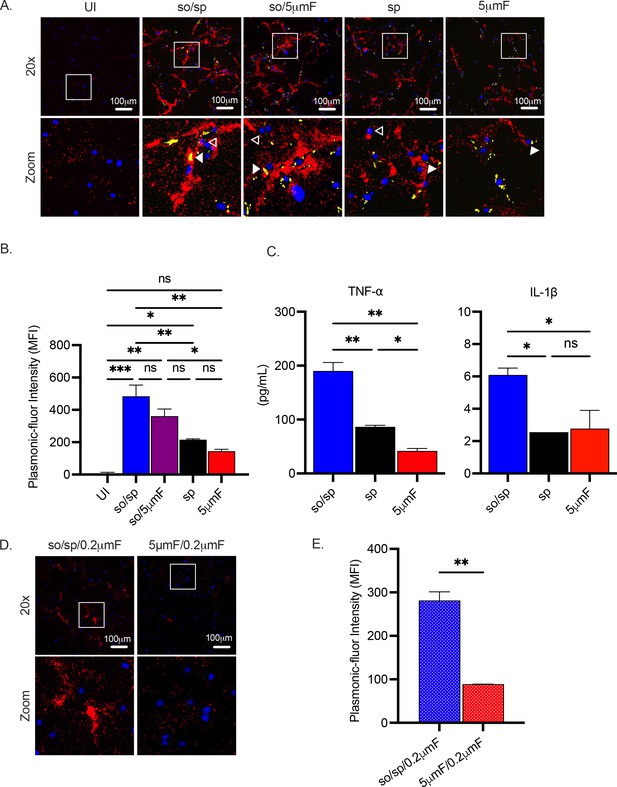
Sonicated bacteria elicit elevated TNF-α and IL-1β secretion.
(A) Using the FluoroDOT assay, BMDMs were grown on a glass bottom plate that was coated with TNF-α capture antibody, infected at an MOI of 10 with H37Rv-GFP prepared by the indicated method, and examined by epifluorescence microscopy (20 X) 6 hpi. Images show Plasmonic-fluor 650 (red), Mtb (GFP), and DAPI (blue). Boxed areas in the image are enlarged in the bottom images. Secretion from infected BMDMs or uninfected bystander cells are highlighted by open or closed white arrowheads, respectively. (B) Data show the quantification of the mean fluorescence intensity (MFI) of the plasmonic-fluor in the entire well from each different condition shown in (A), with statistical significance determined with one-way ANOVA using Tukey’s multiple comparisons test. (C) IL-1β and TNF-α were measured 24 hpi in the culture supernatant of uninfected or Mtb-infected BMDMs (MOI 10) by ELISA. Data shown are mean +/-SD from one representative experiment with three biological replicates per group and two technical replicates per sample. Significance was determined using one-way ANOVA with Tukeys’ multiple comparisons test. (D) Using the FluoroDOT assay, BMDMs grown on a glass bottom plate that was coated with TNF-α capture antibody were exposed to the sterile filtrate of bacterial single cell suspension prepared by either so/sp or 5μmF and examined by epifluorescence microscopy (20 X) 6 hpi. Images show Plasmonic-fluor 650 (red) and DAPI (blue). Boxed areas in the image are enlarged in the bottom images. (E) Data show the quantification of the mean fluorescence intensity (MFI) of the plasmonic-fluor in the entire well from each condition shown in D, with statistical significance determined using an unpaired T test. (A–E) Error bars indicate mean +/-SD. ns not significant; *p<0.05; **<0.01; ***<0.001.
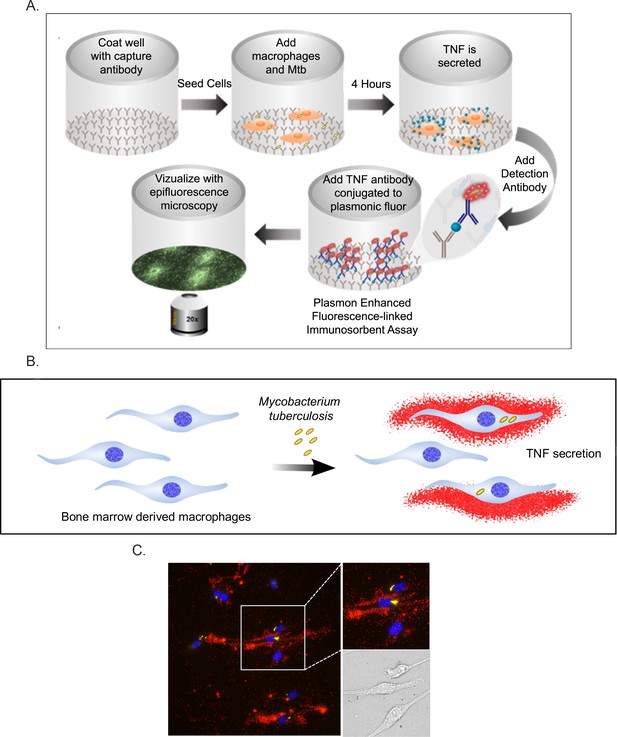
Schematic representation of the FluoroDOT assay.
(A) 96-well glass bottom plates are coated with TNF-α capture antibody, followed by the addition of BMDMs. The macrophages are then infected with Mtb. TNF-α that is secreted by the BMDMs can be bound by the capture antibody. Samples are fixed and then the detection antibody, which is conjugated to plasmon-fluor 650, is added and the plate is visualized using epifluorescence microscopy. (B) Illustration indicating how the assay can reveal TNF-α secretion from Mtb-infected or bystander macrophages. (C) Representative fluorescent image with zoomed region and corresponding brightfield image. The three cells in the images demonstrate examples of an infected macrophage secreting substantial TNF-a (middle), an infected cell with little TNF-α secretion (top), and an uninfected cell with minimal TNF-α secretion (bottom). Macrophages were stained with DAPI (blue); TNF-α is red.
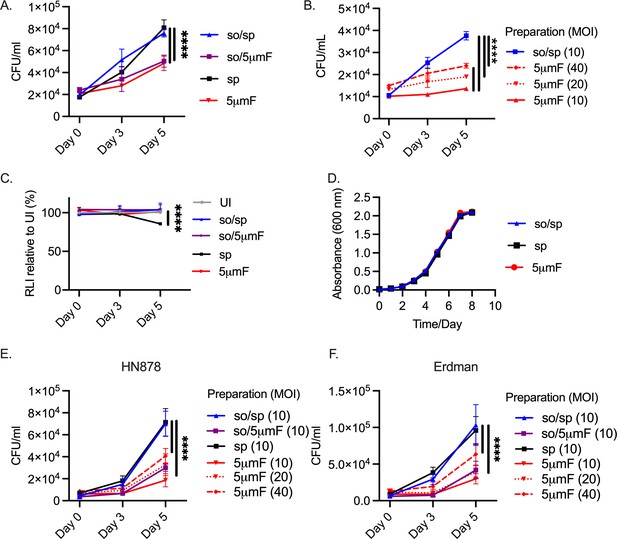
Filtered Mtb are attenuated in BMDMs.
(A) BMDMs were infected with different preparations of Mtb (H37Rv) at an MOI of 10 and intracellular bacteria were enumerated by colony forming units (CFU) 4 hpi, 3 dpi, or 5 dpi. (B) BMDMs were infected with different preparations of Mtb (H37Rv) at an MOI of 10–40 and intracellular bacteria were enumerated by CFU 4 hpi, 3 dpi, or 5 dpi. (C) BMDMs were infected with different preparations of Mtb (H37Rv) at MOI of 10, and BMDM viability was measured using the CellTiter-Glo assay at 4 hpi, 3 dpi, and 5 dpi. Statistical significance between preparations was determined with two-way ANOVA using Dunnett’s multiple comparisons test with selected significance values presented for 5 dpi relative to UI BMDMs. (D) Growth curve of different bacterial H37Rv preparation in liquid media (7H9 media supplemented with 10% Middlebrook OADC, 0.05% Tyloxapol, and 0.2% glycerol). (E–F) BMDMs were infected with different preparations of HN878 (E) or Erdman (F) strains at an MOI of 10–40 and intracellular bacteria were enumerated by CFU 4 hpi, 3 dpi, or 5 dpi. (A–C, E–F) For all CFU and macrophage viability studies, six biological replicates were used per group. Statistical significance was determined for each preparation at 5 dpi by comparing to CFU from BMDMs infected with spin-prepared Mtb using two-way ANOVA with Dunnett’s multiple comparisons test. Selected significance values are presented at 5 dpi. (A–F) Error bars indicate mean +/-SD. ns not significant; ****<0.0001.
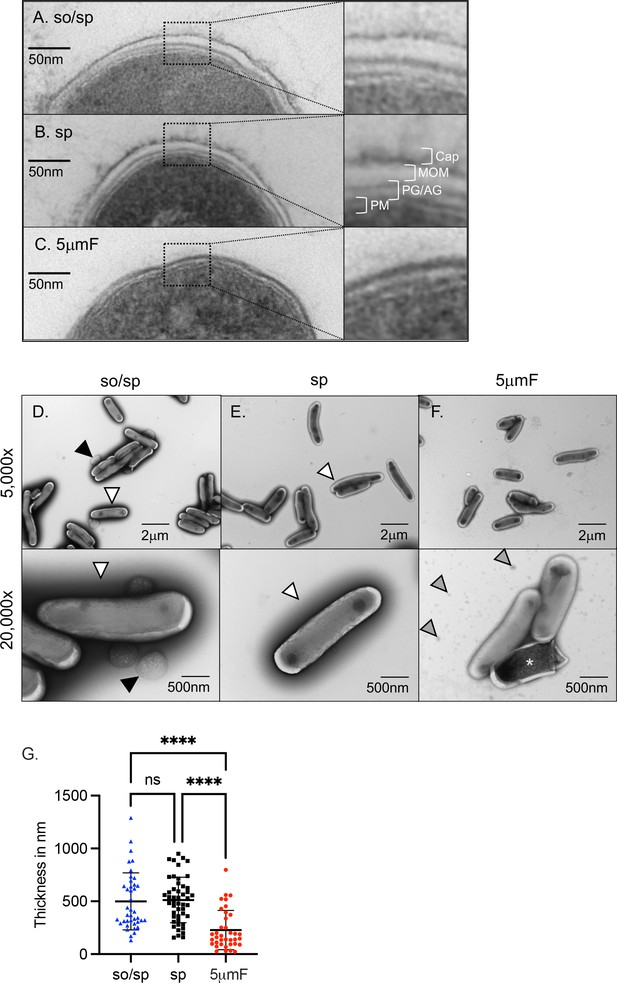
Sonication and filtering affect the bacterial cell wall.
(A–C) TEM of ultrathin cross-sections of Mtb at ×50,000 magnification (left) beside enlarged cross-section of the envelope (right). The plasma membrane (PM), peptidoglycan/arabinogalactan layer (PG/AM), mycobacterial outer membrane (MOM), and capsular layer (Cap) are indicated. (D–F) Mtb were absorbed on freshly glow discharged formvar/carbon-coated copper grids followed by negative staining with 1% aqueous uranyl acetate. Representative images are 5000 x (above) and 20,000 x (below). So/sp-prepared Mtb had round protuberances that were on or near their envelopes indicated by black arrows. Electron-dense outer halos seen surrounding so/sp- and sp-prepared bacteria are indicated with white arrows. Debris seen in the extracellular space of 5μmF-prepared Mtb is indicated with gray arrows. (G) Capsule thickness was measured in nanometers using TEM images from bacteria stained with 1% uranyl acetate. Thickness measurements were compared between preparations with one-way ANOVA using Tukey’s multiple comparisons test. Error bars indicate mean +/-SD. ns not significant; ****<0.0001.
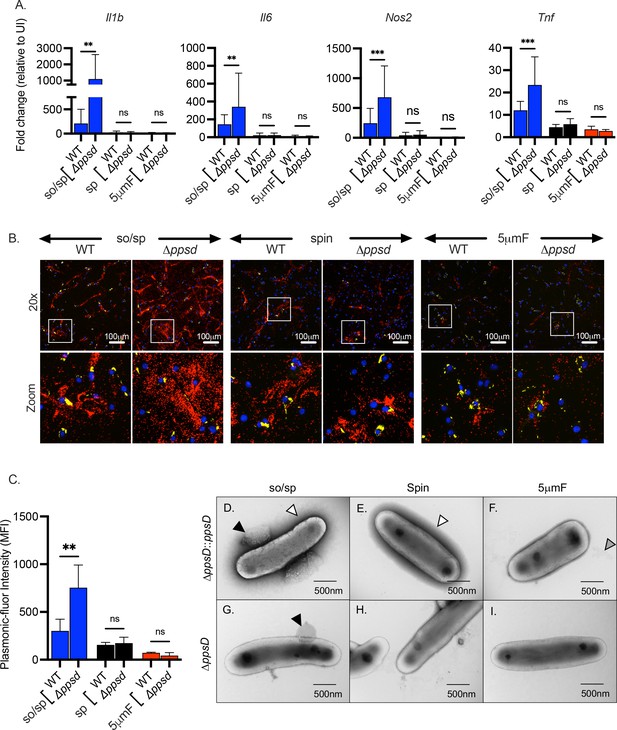
The role of PDIM in inflammatory responses depends upon preparation method.
(A) BMDMs were uninfected or infected with indicated strains of Mtb at an MOI of 10, and gene expression was analyzed by qPCR at 6 hpi. Data are presented as fold change in gene expression relative to uninfected BMDMs of the same mouse genotype. Statistical significance was determined with two-way ANOVA using Tukey’s multiple comparisons test. Data are combined from two to three experiments, each with three biological replicates per group and two technical replicates per sample. (B) Using the FluoroDOT assay, BMDMs were grown on a glass bottom plate that was coated with TNF-α capture antibody, infected at an MOI of 10 with H37Rv-GFP or ∆ppsD-GFP prepared by the indicated method, and examined by epifluorescence microscopy (×20) 6 hpi. Images show Plasmonic-fluor 650 (red), Mtb (GFP), and DAPI (blue). Boxed areas in the image are enlarged in the bottom images. (C) Data show the quantification of the mean fluorescence intensity (MFI) of the plasmonic-fluor in the entire well from each conditions shown in B with statistical significance determined with two-way ANOVA using Tukey’s multiple comparisons test. (D) Bacteria were imaged by allowing indicated Mtb strains to absorb on freshly glow discharged formvar/carbon-coated copper grids followed by negative staining with 1% aqueous uranyl acetate. Round protuberances seen on or near the envelopes of so/sp-prepared H37Rv Mtb are indicated by black arrows, the electron-dense outer halos seen surrounding so/sp- and sp-prepared H37Rv Mtb are indicated with white arrows, and the debris seen in 5μmF-prepared H37Rv Mtb are indicated with gray arrows. (A, C) Error bars indicate mean +/-SD. ns not significant; **p<0.01; ***<0.001.
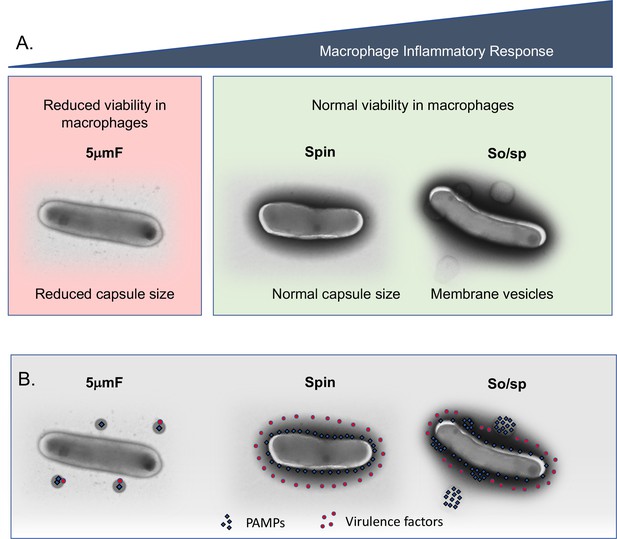
Summary of findings and model describing the impact of bacterial preparation methods on host-pathogen interactions.
(A) Filtered (5μmF), spun , and sonicated (So/sp) Mtb differ in appearance and elicit different macrophage responses. 5μmF bacteria exhibit reduced growth in macrophages and have reduced capsular staining. Bacilli prepared with sonication elicit the strongest inflammatory response and have membrane vesicles and cell envelope protrusions. (B) The findings in (A) can be explained by the following model: filtration disrupts the cell envelope, dispersing and inactivating PAMPs and virulence factors, resulting in reduced macrophage inflammatory responses and reduced intracellular growth. Bacteria that are spun have an intact cell envelope that shields PAMPs and contains virulence factors. Sonication disrupts the cell envelope such that PAMPs are more highly exposed, resulting in increased inflammatory responses; virulence factors remain intact enabling normal growth in macrophages.
Additional files
-
Supplementary file 1
Table of macrophage genes differentially expressed based upon preparation method.
This file lists the genes that were differentially expressed between uninfected macrophages, macrophages infected with Mtb prepared by sonication followed by low-speed spin (so/sp), and macrophages infected with Mtb prepared by passing through a 5 μm filter (5μmF). Infectious were carried out at an MOI of 5 and analyzed at 72 hpi.
- https://cdn.elifesciences.org/articles/85416/elife-85416-supp1-v2.xlsx
-
Supplementary file 2
PCR primers used.
- https://cdn.elifesciences.org/articles/85416/elife-85416-supp2-v2.xlsx
-
Supplementary file 3
Literature review of methods used to generate single cell Mtb suspensions.
(A) Approach used to analyze the literature to define the frequency with which distinct single cell preparation methods are used and how often they are reported. (B) Graph demonstrates the distribution of methods reported. Since some studies used multiple methods, the total does not equal 100.
- https://cdn.elifesciences.org/articles/85416/elife-85416-supp3-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85416/elife-85416-mdarchecklist1-v2.docx





