High-throughput mapping of single-neuron projection and molecular features by retrograde barcoded labeling
Figures

MERGE-seq characterized single-neuron transcriptomes and projectomes simultaneously.
(A) Schematic diagram of the experimental workflow. (B) rAAV2 plasmid vector design, and schematic of designed primers to recover cell barcode and UMI in read 1, and 3’ tail of EGFP and virus barcode in read 2. According to the recommendation of 10x Genomics, a faithful mapping should cover 28 bp for read 1 and 91 bp for read 2. In our design, 150 bp pair-end sequencing can sufficiently meet the need to recover cell barcode, UMI and virus barcode. (C) Umap embedding of transcriptional clustering results for all vmPFC cells. (D) Stacked violin plots showing the expression of markers for each cluster. (E) Heatmap showing the gene-expression correlation between major cell types defined by scRNA-seq of this study and Bhattacherjee et al., 2019. (F) Umap embedding of all determined barcoded cells labeled in blue. (G) Bar plot showing frequency of barcoded (blue) and non-barcoded (grey) cells in all recovered cell types. In (C–E), 21,261cells were represented. In (F, G), 20,047cells were represented. 1214 cells with exceptionally high nUMIs were removed.
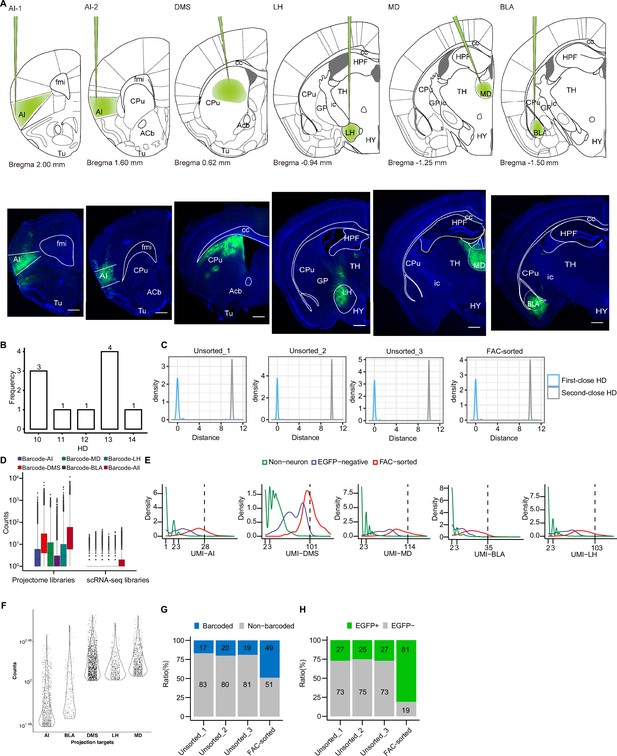
Validation of rAAV2-retro injection sites and determination of valid barcoded cells.
(A) The position of injection sites (AI-1, AI-2, DMS, LH, MD, BLA) to deliver rAAV2-retro-EGFP plotted on coronal section diagrams (top). The left corner values indicate the anteroposterior distance of the section from bregma. Representative immunohistochemistry images showing rAAV2-retro-EGFP injection sites in coronal sections (bottom). Scale bars: 500 μm. AI-1, agranular insular cortex, anterior; AI-2, agranular insular cortex, posterior. (B) Frequency of hamming distance (HD) of five reference barcode sequences. (C) Density distribution of HD of best and second-best hit when comparing barcode sequences form reads to and five barcode references. The distance to the best hit (identified barcode) is 0 across four samples as we used perfect match for the identification. And the distance of the second best hit is ~10, showing that there are sufficient sequence difference to identify the right barcode among five references during sequencing. (D) Boxplot displaying the count distribution of UMIs for neurons with projections identified by different barcodes and a sum of all five barcodes across projectome and scRNA-seq libraries. (E) Density plots contrasting the distribution of UMIs for each target region between non-neuronal cells (green), EGFP-negative cells (blue), and FAC-sorted cells (red). The dashed line indicates a threshold for UMI counts of a barcoded neuron. Note that EGFP-negative cells are subset of FAC-sorted group and determined as nUMI of EFGP RNA = 0. (F) Violin plots of Log10 normalized projection index barcode counts. Note that after choosing the UMI counts threshold, UMI counts below threshold were dropped to zero. (G) Stacked bar plot showing barcoded and non-barcoded cell ratio in FAC-sorted group or unsorted group as determined by stringent UMI threshold. (H) Stacked bar plot showing EGFP-positive (nUMI of EFGP RNA >0) and EGFP-negative cells (nUMI of EFGP RNA = 0) ratio in FAC-sorted group or unsorted group as determined by scRNA-seq.
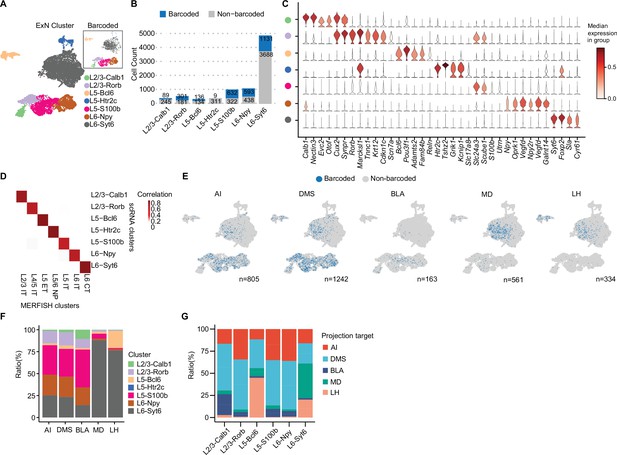
MERGE-seq unravels transcriptomic heterogeneity of projection target-defined vmPFC neurons.
(A) Umap embedding of excitatory neuron subtype annotation. (B) Bar plot showing frequency of barcoded (blue) and non-barcoded (grey) neurons in distinct neuron subtypes. (C) Stacked violin plot showing the expression of markers for each neuronal subtype. (D) Heatmap showing the gene-expression correlation between excitatory subtypes defined by Multiplexed Error-Robust Fluorescence in situ Hybridization (MERFISH) and scRNA-seq. MERFISH data were downloaded from Bhattacherjee et al., 2023. (E) Umap embeddings of barcoded (blue) neurons projecting to each target. Number indicates the number of barcoded cells for each target. (F) Bar plot describing the distribution of neuronal subtypes for barcoded neurons associated with each projection target. Neuronal subtype color codes are the same as in (A), number of barcoded cells are same as the number indicated in (E) for each target. (G) Bar plot describing the distribution of projection targets for barcoded neurons associated with each neuronal type. In (A, C, D), 9368 cells in total were represented. In (B, E, F), 8210 cells in total were represented. In (G), cell numbers represented are as follows: L2/3-Calb1=72 cells, L2/3-Rorb=331 cells, L5-Bcl6=145 cells, L5-S100b=766 cells, L6-Npy=526 cells, L6-Syt6=1264 cells.
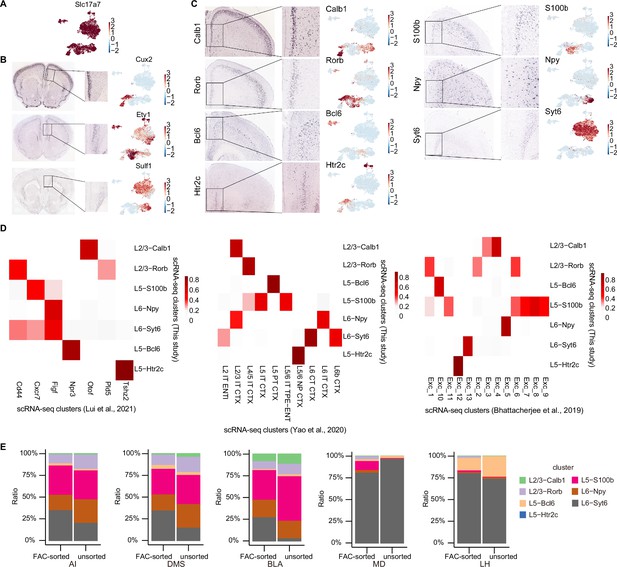
Layer and cluster annotation using the mouse brain atlas and published scRNA-seq transcriptomes, and projection patterns per mouse.
(A) Normalized Slc17a7 (vGlut1) expression for all extracted excitatory neurons. (B) In situ hybridization of typical layer-specific markers within the vmPFC region from the Adult Mouse Brain Atlas. Cux2 is layer 2/3-specific; Etv1 is layer 5-specific; Sulf1 is layer 6-specific (left). Normalized expression of Cux2, Etv1, and Sulf1 at umap embedding (right). (C) In situ hybridization of typical neuronal subtype markers in the vmPFC from the Adult Mouse Brain Atlas. Calb1 and Rorb are layer 2/3-specific; Htr2c and S100b are layer 5-specific; Bcl6 is around the transition of layer 2/3 and layer 5. Syt6 and Npy are layer 6-specific, though Npy is distributed sporadically. Corresponding normalized gene expression embedded in umap is plotted in the right panel. (D) Heatmap showing the gene-expression correlation between excitatory subtypes defined by scRNA-seq of this study and (Lui et al., 2021), (left) or (Yao et al., 2021), (middle) or (Bhattacherjee et al., 2019), (right). (E) Stacked bar plots showing neuronal subtype composition of pooled unsorted mice and pooled FAC-sorted mice for each projection target. Statistical approach was not applied due to the limitations of having a single observation per cluster per group.
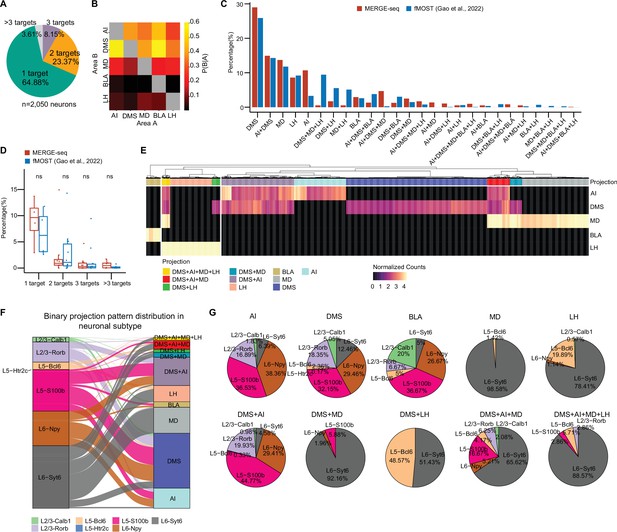
MERGE-seq reveals projection diversity within the vmPFC.
(A) Pie chart indicating the number of projection targets for barcoded vmPFC neurons recovered by MERGE-seq. (B) Heatmap showing the probability that a neuron projecting to area A also projects to area B. (C) Bar graph illustrating the percentage of neuronal projection pattern of all projection patterns given five projection targets inferred by MERGE-seq (red bars) versus the 1155 fMOST-based single-neuron projectome data (blue bars) (Gao et al., 2022). (D) Boxplot comparison of percentage of neurons with different projection targets identified by MERGE-seq and fMOST. (E) Heatmap showing normalized projection strength. Rows represent the projection targets and columns represent the cells labeled by the top 10 binary projection patterns or labeled by transcriptional neuron subtypes. (F) Alluvial plot showing the 10 most frequent projection patterns distribution into neuronal subtypes. (G) Pie charts describing the projection patterns from (E) partitioned by neuronal subtype. In (A, B), 2050 barcoded neurons were represented. In (C, D), 2050 barcoded neurons from MERGE-seq data were represented, 1155 cells with fMOST data were represented (Gao et al., 2022). In (E–G), 1853 barcoded neurons (top 10 frequent projection patterns) were represented.
-
Figure 3—source data 1
Related to Figure 3C, D, F and G.
Files contain raw data for Figure 3C and D, statistical summary for Figure 3D, raw data for Figure 3F and G.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig3-data1-v2.xlsx
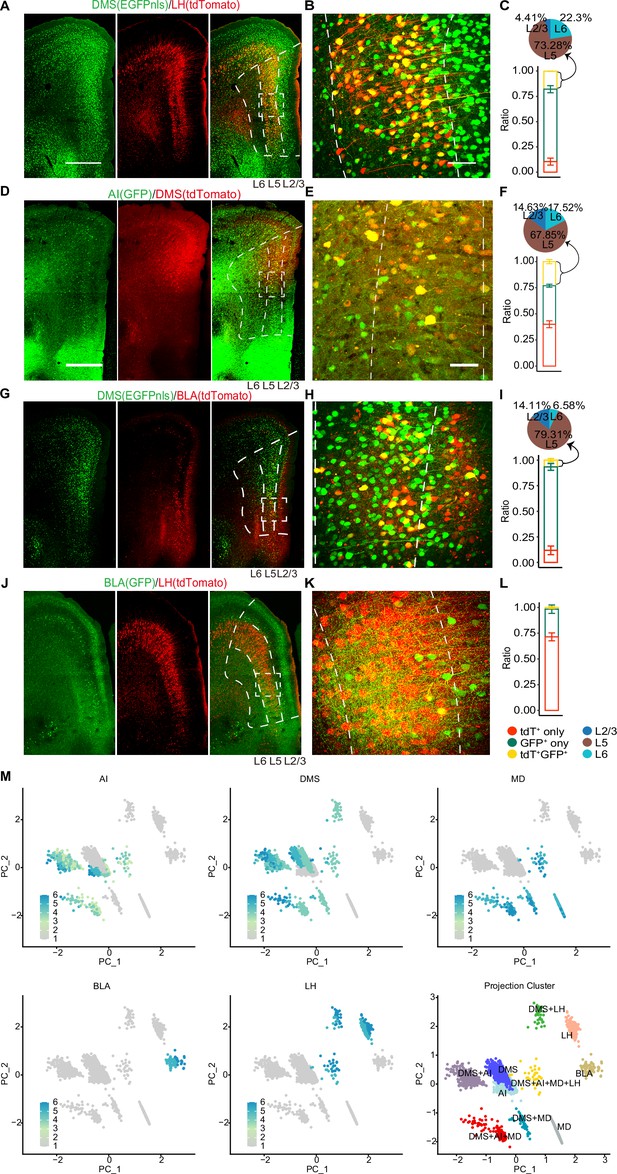
Immunostaining of dual-color, retrogradely labeled neurons and quantification, PCA plot of projection clusters.
(A–L) Immunostaining of dual-color traced retrograde labeled neurons of selected targets DMS (GFPnls) /LH (tdTomato), AI (GFP) /DMS (tdTomato), DMS (GFPnls) /BLA (tdTomato), and BLA (GFP) /LH (tdTomato). (A, D, G, J). Dotted line depicts layers 2/3, 5, and 6 of the vmPFC. Scale bars, 500 µm. (B, E, H, K) Enlarged view of the dotted box in (A, D, G, J). Scale bars, 100 µm. (C, F, I, L) Histogram shows quantitative data for single- (red, green) and double- (yellow) labeled neurons as mean percentages of total rAAV2-retro labeled neurons (n=3 mice). Data are presented as mean ± SD. Pie chart showing layer distribution of double (yellow) labeled neurons. (M) Normalized projection index barcode expression on PC1 and PC2 embeddings and binary projection annotation labeled on PC1 and PC2 embeddings. Note that only the 10 most frequent binary projection patterns were included. Data are the mean ± SD.

Transcriptional profiling of projection target-specific vmPFC neurons.
(A) Volcano plots DEGs of MD-projecting versus non-MD-projecting vmPFC neurons. Assigned DEGs (red dots) were determined using threshold: Log2 fold change = 0.5, p value cutoff=10–10. (B) Immunostaining of EGFP (MD) and tdTomato (LH), and RNA FISH of Syt6. (i, ii) Enlarged view of dotted box in (B). (i) represents typical view at layer 6 and (ii) represents typical view at layer 5. Arrow head indicates Syt6+EGFP+ neurons. (C) Quantifications of (B). (B) Scale bars, 200 μm. i, ii in (B) Scale bars, 50 μm. N=3 mice. Data are presented as mean ± SD. In (A), 8210 cells were represented.
-
Figure 4—source data 1
Related to Figure 4A, Figure 4—figure supplement 1.
Files contain the raw differentially differentially expressed genes (DEGs) list used for plotting volcano plots. For each target, DEGs were calculated using target-barcoded cells against non-target-barcoded cells (e.g., AI barcoded versus non-AI-barcoded, DMS barcoded versus non-DMS-barcoded, etc.). MAST algorithm was used to do DE testing.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Related to Figure 4B and C.
Files contain the raw quantitative data of immunostaining results.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig4-data2-v2.xlsx
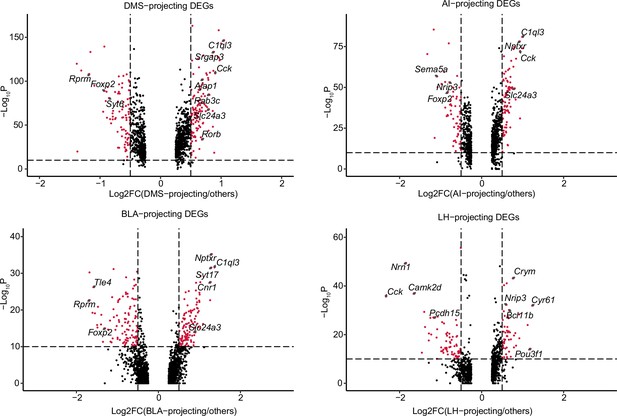
Transcriptional profiling of projection target-specific vmPFC neurons.
Volcano plots DEGs of DMS-projecting versus non-DMS-projecting vmPFC neurons, AI-projecting versus non-AI-projecting vmPFC neurons, BLA-projecting versus non-BLA-projecting vmPFC neurons, and LH-projecting versus non-LH-projecting vmPFC neurons. Assigned DEGs (red dots) were determined using threshold: Log2 fold change = 0.5, p value cutoff=10–10.
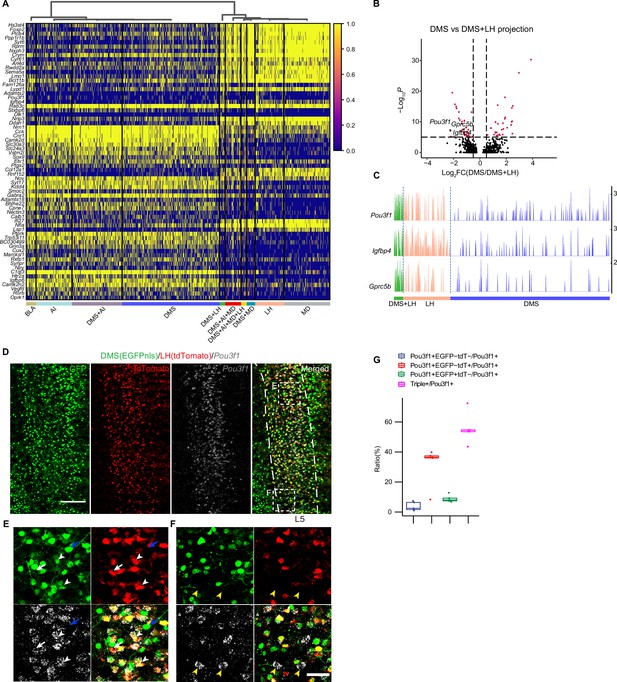
Molecular features of single vmPFC neuron with collateral projections to downstream targets.
(A) Heatmap showing scaled expression of calculated DEGs based on 10 projection patterns. Top 10 DEGs ordered by average log2 fold change of each pattern were selected. (B) Volcano plot showing genes differentially expressed in the DMS + LH-bifurcated projection pattern compared to the DMS-dedicated projection pattern. (C) Track plots showing normalized data of the selected DEGs in DMS-dedicated, LH-dedicated, and DMS + LH-bifurcated projection pattern. (D–F) Examining Pou3f1 and DMS + LH-bifurcated projection pattern using RNA FISH and immunostaining of dual-color traced retrograde labeled neurons. Virus injection scheme was the same as in Figure 3—figure supplement 1. Scale bars, 200 µm. (E, F) Enlarged view of dotted box in (D). Arrow heads indicate Pou3f1+EGFP+tdTomato+ neurons, white arrows indicate Pou3f1+EGFP-tdTomato+ neurons, blue arrows indicate Pou3f1-EGFP+tdTomato- neurons, and yellow arrowheads indicate Pou3f1+EGFP-tdTomato- neurons. Scale bars, 50 µm. (G) Quantification of (D). N=3 mice, Data are presented as mean ± SD. In (A), 1,853 barcoded neurons (top 10 frequent projection patterns) were represented. In (C), 805 barcoded neurons (projection pattern DMS + LH = 35, LH = 176, DMS = 594) were represented.
-
Figure 5—source data 1
Related to Figure 5B.
Files contain the raw differentially differentially expressed genes (DEGs) list used for plotting volcano plots. DEGs were calculated using DMS-barcoded cells against DMS + LH-barcoded cells. MAST algorithm was used to do DE testing.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Related to Figure 5D–G.
Files contain the raw quantitative data of immunostaining results.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig5-data2-v2.xlsx
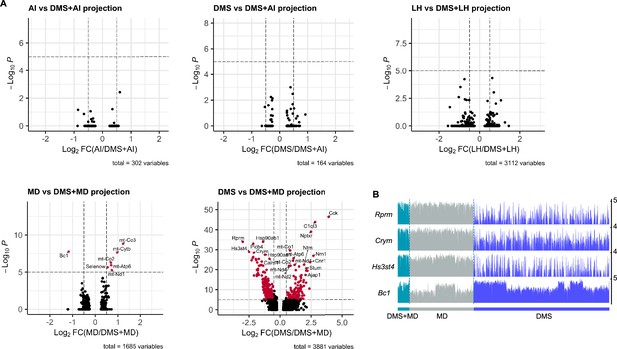
DEGs between dedicated projection neurons versus bifurcated neurons.
(A) Volcano plots of DEGs calculated between A and A/B projection patterns. See also Figure 5. Assigned DEGs (red dots) were determined using threshold: Log2 fold change = 0.5, p value cutoff=10–5. (B) Track plots showing normalized data of the selected DEGs (DMS versus DMS + MD projection) in DMS-dedicated, MD-dedicated, and DMS + MD-bifurcated vmPFC neurons.
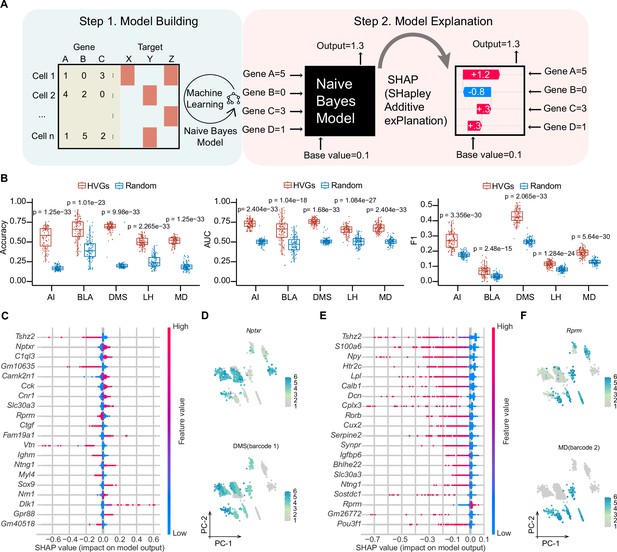
Machine learning-based modeling predicts projection patterns based on gene expression.
(A) Schematics of machine learning modeling steps. (B) Prediction accuracy (left panel), AUC score (middle panel) and F1 score (right panel) of top HVGs and random chosen equal number of genes for modeling building. A total of 100 trials have been performed by randomly sampling 1000 cells from 8210 cells. Top 50 HVGs or 50 randomly chosen genes were used as features per trial. Comparisons were made between models built by the HVGs group and random genes group for each projection target. The displayed p value was computed using a two-sided Wilcoxon test. Data are the mean ± SD. (C, E) SHAP summary plots of DMS and MD showing important features (genes) with feature effects. For each model, non-barcoded cells were encoded to class 0 and barcoded cells were encoded to class 1. Models were built using top 50 HVGs. (D, F) Normalized expression of the most important genes with positive feature effects in Naïve Bayes modeling of DMS (D) or MD (F) and normalized expression of barcode 1 representing DMS-projecting (E) or barcode 2 representing MD-projecting (F) on PC1 and PC2 embeddings. Note that bottom panel of (D, F) is identical to DMS and MD barcode expression in Figure 3—figure supplement 1M. In (D, F), 1853 barcoded neurons (top 10 frequent projection patterns) were represented. In (C, E), For calculating SHAP values, both the training and testing datasets were subsampled to include 1500 cells each.
-
Figure 6—source data 1
Related to Figure 6B, Figure 6—figure supplement 1B.
Files contain raw data and statistical summary, raw data and for Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/85419/elife-85419-fig6-data1-v2.xlsx
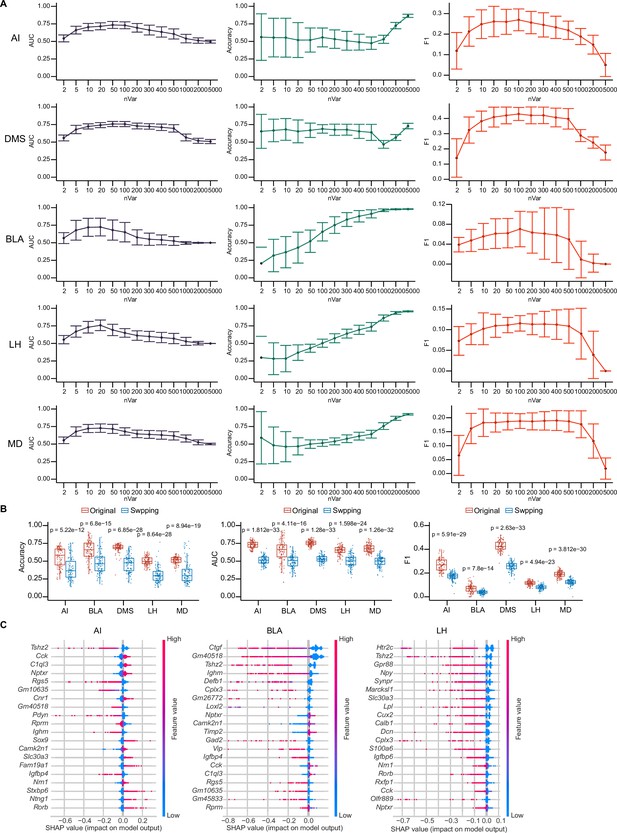
SHAP summary plots of Naïve Bayes models.
(A) Prediction accuracy (left panel), AUC score (middle panel) and F1 score (right panel) by tuning number of HVGs used for naïve bayes modeling building. A total of 100 trials have been performed by randomly sampling 1000 cells from 8210 cells and calculating top HVGs per trial. (B) Prediction accuracy (left panel), AUC score (middle panel) and F1 score (right panel) of original top 50 HVGs and after swapping barcoded/non-barcoded cell labels for modeling building. A total of 100 trials have been performed by randomly sampling 1000 cells from 8210 cells. In each trial, barcoded/non-barcoded cell labels were swapped. The number of swapped cells depends on the minimum number of barcoded cells or non-barcoded cells. Models were built with the top 50 HVGs using original labels (Original) or labels swapped (Swapping) for comparison for each projection target. The displayed p value was computed using a two-sided Wilcoxon test. Data are the mean ± SD. (C) SHAP summary plots of AI, BLA, and LH showing important features (genes) with feature effects. For each model, non-barcoded cells were encoded to class 0 and barcoded cells were encoded to class 1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-GFP (Rat Monoclonal) | Nacalai | Cat# 04404–84, RRID:AB_10013361 | IHC(1:500) |
| Antibody | Anti-tdTomato (Goat Polyclonal) | OriGene | Cat#AB8181-200, RRID:AB_2722750 | IHC(1:500) |
| Antibody | Hoechst 33342 | Lifetech | Cat#H3570 | IHC(1:1000) |
| Antibody | Donkey anti-rat Alexa Fluor 488 (Donkey Polyclonal) | Invitrogen | Cat#A21208 | IHC(1:800) |
| Antibody | Donkey anti-goat Alexa Fluor 568 (Donkey Polyclonal) | Invitrogen | Cat#A11057 | IHC(1:800) |
| Recombinant DNA reagent | pAAV-CAG-tdTomato (plasmid) | Addgene | Cat#59462 | |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-0-SV40 polyA (plasmid) | This paper | Cat#190864 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-1-SV40 polyA (plasmid) | This paper | Cat#190865 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-2-SV40 polyA (plasmid) | This paper | Cat#190866 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-3-SV40 polyA (plasmid) | This paper | Cat#190867 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-4-SV40 polyA (plasmid) | This paper | Cat#190868 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-5-SV40 polyA (plasmid) | This paper | Cat#190869 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-6-SV40 polyA (plasmid) | This paper | Cat#190870 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-7-SV40 polyA (plasmid) | This paper | Cat#190871 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-8-SV40 polyA (plasmid) | This paper | Cat#190872 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-9-SV40 polyA (plasmid) | This paper | Cat#190873 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFP barcode-10-SV40 polyA (plasmid) | This paper | Cat#190874 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFPnls barcode-206-SV40 polyA (plasmid) | This paper | Cat#190875 | Submitted to Addgene |
| Recombinant DNA reagent | pAAV-CAG-EGFPnls barcode-210-SV40 polyA (plasmid) | This paper | Cat#190876 | Submitted to Addgene |
| Chemical compound, drug | AMPA receptor antagonist CNQX | Abcam | Cat#ab120017 | working concentration:10 µM |
| Chemical compound, drug | NMDA receptor antagonist D-AP5 | Abcam | Cat#ab120003 | working concentration:50 µM |
| Chemical compound, drug | 2-Mercaptoethanol | Sigma | Cat#M6250 | working concentration:0.067 mM |
| Chemical compound, drug | EDTA | Invitrogen | Cat#15575020 | working concentration:1.1 mM |
| Chemical compound, drug | L-Cysteine hydrochloride monohydrate | Sigma | Cat#C7880 | working concentration:5.5 mM |
| Chemical compound, drug | Deoxyribonuclease I | Sigma | Cat#D4527 | working concentration:100 units/ml |
| Chemical compound, drug | Protease | Sigma | Cat#P5147 | working concentration:1 mg/ml |
| Chemical compound, drug | Dispase | Worthington | Cat#LS02106 | working concentration:1 mg/ml |
| Chemical compound, drug | Papain | Worthington | Cat#LS003126 | working concentration:20 units/ml |
| Commercial assay or kit | Debris Removal Solution | Miltenyi | Cat#130-109-398 | |
| Commercial assay or kit | Chromium Single Cell 3' Reagent Kits (v3) | 10 X Genomics | Cat#PN1000075 | |
| Commercial assay or kit | NEBNext Ultra II Q5 Master Mix | NEB | Cat#M0544L | |
| Commercial assay or kit | SPRIselect | Beckman | Cat#B23317 | |
| Commercial assay or kit | Mm-Syt6 | ACD Bioscience | Cat#449641 | |
| Commercial assay or kit | Mm-Pou3f1-C2 | ACD Bioscience | Cat#436421-C2 | |
| Sequence-based reagent | P5-Read1 | This paper | PCR primers | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC |
| Sequence-based reagent | P7-index-Read2-EGFP | This paper | PCR primers | CAAGCAGAAGACGGCATACGAGATAGGATTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGgCATGGACGAGCTGTACAAG |
Additional files
-
Supplementary file 1
Marker genes of 2nd round clustering of “Excitatory neuron” cluster, related to Figure 2.
The inserted umap shows the number of UMI counts (nCount_RNA) per cluster, number of genes (nFeature_RNA) per cluster, percentage of mitochondria genes (percent_mt) per cluster.
- https://cdn.elifesciences.org/articles/85419/elife-85419-supp1-v2.xlsx
-
Supplementary file 2
Marker genes of 7 scRNA-seq clusters from all excitatory neurons, related to Figure 2.
Table with marker genes for each cluster calculated using Seurat package using Wilcoxon test.
- https://cdn.elifesciences.org/articles/85419/elife-85419-supp2-v2.csv
-
Supplementary file 3
Marker genes of 8 clusters from all cells, related to Figure 1.
Table with marker genes for each cluster calculated using Seurat package using Wilcoxon test.
- https://cdn.elifesciences.org/articles/85419/elife-85419-supp3-v2.csv
-
Supplementary file 4
Median gene detection metrics for different major cell types, related to Figure 1.
- https://cdn.elifesciences.org/articles/85419/elife-85419-supp4-v2.csv
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85419/elife-85419-mdarchecklist1-v2.docx





