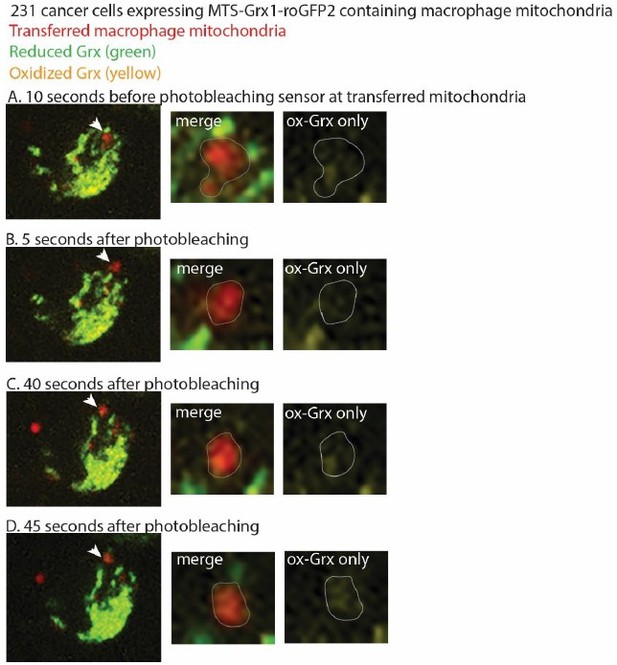
Transferred mitochondria accumulate reactive oxygen species, promoting proliferation
Figures
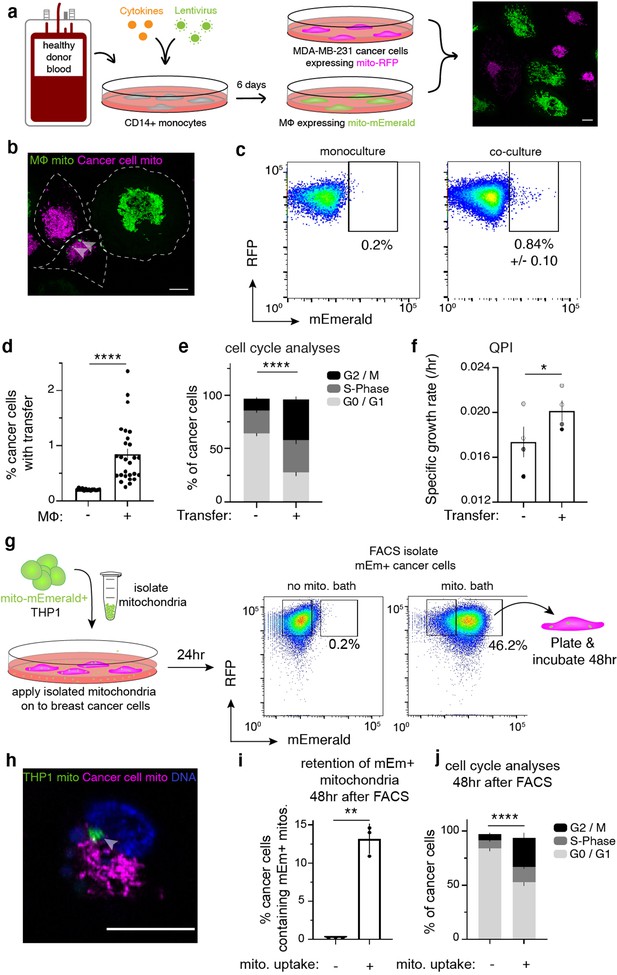
Cell-contact-mediated transfer of macrophage mitochondria leads to increased cancer cell proliferation.
(a) CD14+ monocytes harvested from human blood are transduced and differentiated for 6 days. Mito-mEm +macrophages (green) are co-cultured with MDA-MB-231 cells (231 cells) expressing mito-RFP (magenta; right image). (b) Confocal image showing transferred mitochondria (green, arrowhead) in a 231 cell (magenta, cell outline in white). (c) Representative flow cytometry plots depicting mitochondrial transfer (black box) within a population of co-cultured mito-RFP+ 231 cells (right) compared to monoculture control (left) with background level of mEmerald (mEm) fluorescence set at 0.2%. (d) Aggregate data of mitochondrial transfer rates across macrophage donors. Each data point represents one replicate (N=14 donors). (e) Analysis of proliferative capacity by quantifying Ki-67 levels and DNA content in co-cultured 231 cells after 24 hr. Percentage of cancer cells within a specific cell cycle phase with or without transfer is shown. A significantly different percent of recipient cells occupies G2/M (black) phases of the cell cycle compared to non-recipient cells (N=4 donors; statistics for G2/M only). (f) Co-cultured recipient 231 cells have a significantly higher specific growth rate compared to non-recipients (N=60 cells (control), 115 (recipient) over 4 donors indicated as shades of gray). (g) Schematic of mitochondrial isolation and bath application on MDA-MB-231 cells. Mitochondria are isolated from mito-mEmerald expressing THP-1 monocytes and bath applied at 20–30 µg/mL for 24 hr. Cancer cells which had taken up mEm+ mitochondria are then FACS-isolated and plated for 48 hr for further analyses. (h) Representative confocal image showing mito-RFP-expressing 231 cell (magenta) that had taken up macrophage mitochondria (green, grey arrow). (i) 48 hr after FACS-isolating 231 cells with macrophage mitochondria, flow cytometry was used to determine percent of daughter cells which still contain mEm+ mitochondria. N=3 biological replicates. (j), Cell cycle analysis of daughter cells 48 hr after FACS-isolation of 231 cells that had taken up macrophage mitochondria. N=3 biological replicates. For all panels, standard error of the mean (SEM) is displayed and scale bars are 10 µm. Mann-Whitney (d), two-way ANOVA (e, j), Welch’s t-test (f, i), *p<0.05; **p<0.01; ****p<0.0001.
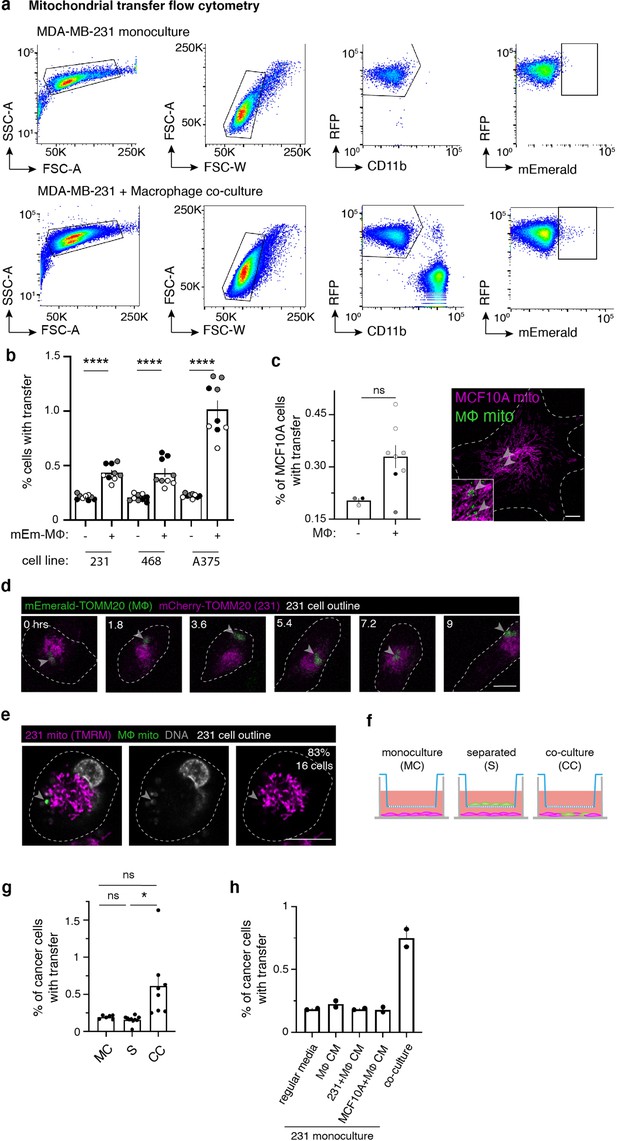
Macrophages transfer mitochondria to cancer cells.
(a) Representative flow cytometry plots of mito-RFP MDA-MB-231 monocultured cells used as a control (top) and mito-RFP 231/mito-mEm macrophage co-cultures after 24 hr (bottom). (b) Panel of cancer cell lines – MDA-MB-231 (‘231’), MDA-MB-468 (‘468’) and A375 – co-cultured with mEm-positive (transduced, ‘+’) or mEm-negative (untransduced, ‘-’) macrophages. Mitochondrial transfer rates determined by flow cytometry, as described in (a). Different donors (N=3) indicated as shades of gray. (c) Left: Bar graph quantifying mitochondrial transfer to MCF10A cells after 24 hr. (N=3 donors indicated by shades of gray). Right: Confocal image showing macrophage mitochondria (green, arrowhead) in a MCF10A cell (magenta). (d) Stills from a time-lapse showing a 231 cell (mCherry-TOMM20, magenta) containing transferred macrophage mitochondria (mEmerald-TOMM20, green, arrowhead). Timepoints indicated in upper left. (e) Single Z-plane of a 231 cell labeled with mitochondrial dye TMRM (magenta) with macrophage mitochondria (green, arrowhead) containing DNA (gray). 83% of transferred mitochondria contain DNA (N=16 cells). (f) Schematic depicting trans-well experiments. 231 cells (magenta) were plated alone (left), separated from macrophages (green; middle), or plated together (right). (g) Percent of cancer cells with transfer in conditions depicted in f, (N=3 donors). (h), Conditioned media (CM) experiments showing percent of cancer cells with transfer when co-cultured in media type listed on the x-axis (N=2 experiments). Each dot represents one replicate for all panels. Error bars represent standard error of the mean (SEM) and scale bars are 10 µm. Two-way ANOVA (b, g), Mann-Whitney test (c), ****p<0.0001.
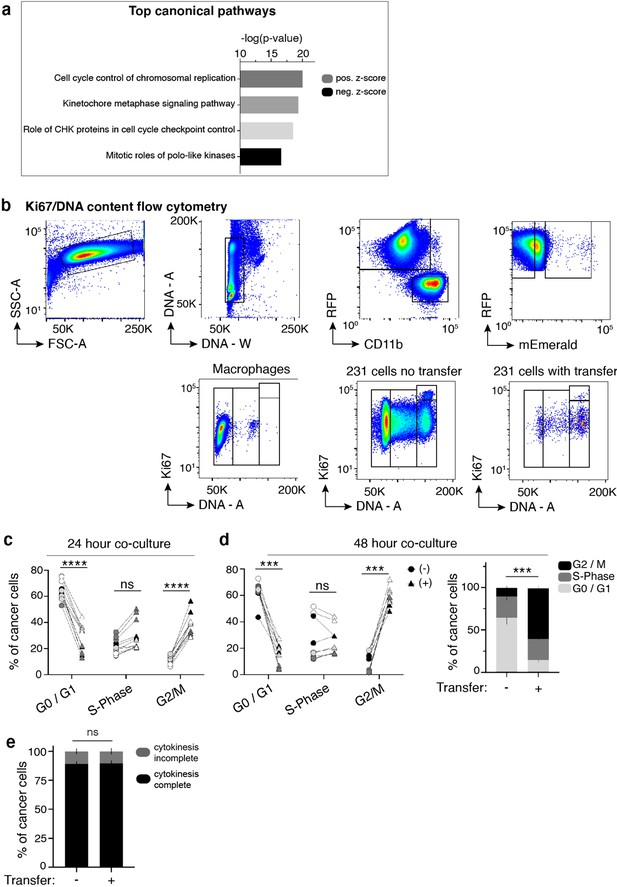
Cancer cells will macrophage mitochondria exhibit increased proliferation.
(a) Cancer cells and macrophages were cocultured for 24 hours and scRNA-seq was performed. Ingenuity Pathway Analysis of scRNA-seq data reveals significant changes in canonical cell proliferation pathways in co-cultured cancer cells that received macrophage mitochondria compared to co-cultured cancer cells that did not. (b) Representative flow cytometry plots of mito-RFP 231/mito-mEm macrophage co-cultures stained for Ki67 and DNA content. (c) 24 hr data from Figure 1e but separated as percent of cancer cells in each phase of the cell cycle with (triangle) or without transfer (circle). (d) 48 hr of co-culture as described in (c) with aggregate data in stacked bar graphs to the right (stats displayed for G2/M only). Each pair in (c) and (d) represents cells within one technical replicate and each donor is represented by color (N = 4 donors for both data sets). (e) Time-lapse imaging was used to quantify the amount of co-cultured 231 cells that did (black) or did not complete (gray) cytokinesis over a 48-hr period (N = 416 cells, 4 donors). Error bars represent SEM. Two-way ANOVA (c–e), ***p<0.001; ****p<0.0001.
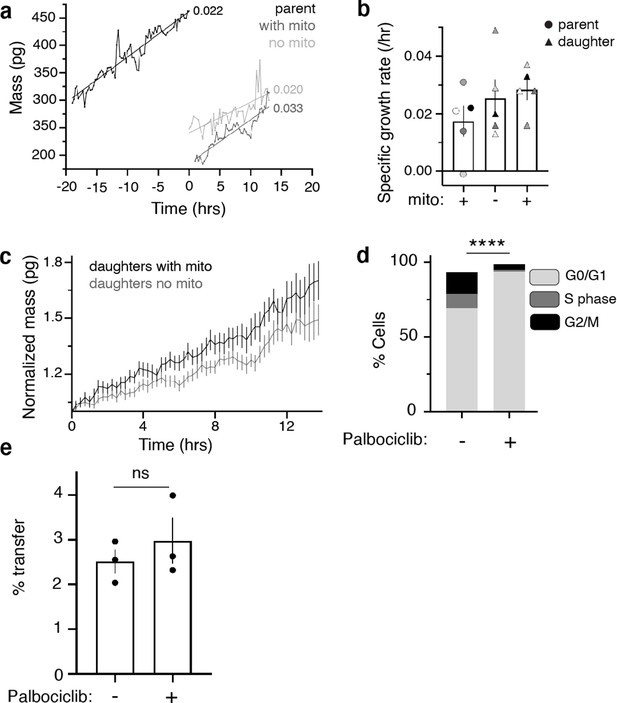
Mitochondrial transfer leads to sustained increased growth rate in daughter cancer cells.
(a) QPI data of mass over time measurements for a single triad consisting of one parent cell (black) and two daughters that inherited (gray) or did not inherit (light gray) the parent’s macrophage mitochondria. Specific growth rate (slope of best fit line normalized by average mass) is listed next to line trace of corresponding cell. (b) QPI data of specific growth rate of 5 individual triads (indicated by color). Parent and daughter cells are indicated with shape (legend on graph). (c) QPI data of normalized mass in picograms (pg) over time in hours (hrs) for daughter cells that did (black) or did not (gray) inherit the parent’s macrophage mitochondria normalized to daughter cell initial mass. Error bars represent SEM. (d) Cell cycle analyses MDA-MB-231/macrophage co-cultures after 24 hr 1 µM Palbociclib treatment. Two-way ANOVA comparing G1/G0 fractions in each condition, ****p<0.0001. When comparing G2/M fractions across conditions **p=0.0017. n=1 in technical triplicate. (e) MDA-MB-231/macrophage cocultures were treated with 10 µM Palbociclib for 24 hr. Mitochondrial transfer rates were determined via flow cytometry as previously described. Unpaired t-test p=0.4675, n=1 in technical triplicate.
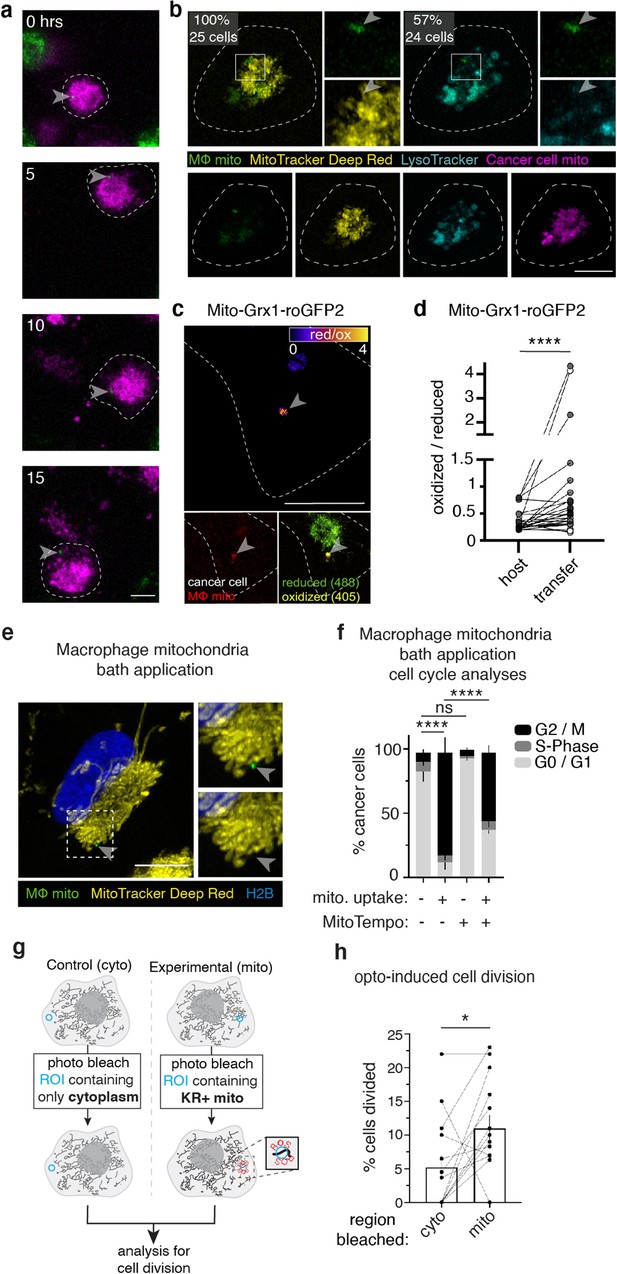
Transferred macrophage mitochondria are long-lived, depolarized, and accumulate reactive oxygen species, promoting cancer cell proliferation.
(a) Stills from time-lapse imaging depicting the longevity of the transferred mitochondria (green, arrowhead) within a 231 cell (magenta, cell outline in white). Time elapsed listed in left corner. (b) Confocal image of a mito-RFP+ 231 cell (magenta) containing macrophage mitochondria (green, arrowhead) stained with MTDR (yellow) and LysoTracker (teal). MTDR does not accumulate in 100% of donated mitochondria (N=25 cells, 5 donors). Majority (57%) of donated mitochondria do not colocalize with LysoTracker signal (N=24 cells, 4 donors). (c) Ratiometric quantification of mito-Grx1-roGFP2 biosensor mapped onto the recipient 231 cell with fire LUT (top panel). Confocal image of mito-Grx1-roGFP2-expressing 231 cell (bottom right, green and yellow) containing a macrophage mitochondria (bottom left, red, arrowhead). (d) Ratiometric measurements of the mito-Grx1-roGFP2 sensor per 231 cell (paired dots) at a region of interest containing the host mitochondrial network (host) or a transferred mitochondria (transfer). Cells were co-cultured for 24 hr (N=27 cells, 3 donors indicated in shades of gray). (e) Exogenous purified macrophage mitochondria (green) is void of mitochondrial membrane potential (MitoTracker Deep Red-negative, yellow, arrowhead) in cancer cells. (f) Cell cycle analysis of cancer cells with exogenous purified macrophage mitochondria versus sister cells that did not take up exogenous purified mitochondria, either treated with vehicle or 100 μM mitoTEMPO (mitochondrially-targeted superoxide scavenger. N=3 donors; statistics for G2/M only). (g) Schematic of optogenetic experiments to generate data in (h). Cells expressing mito-KillerRed are photobleached in a specific ROI containing either cytoplasm only (left) or mito-KillerRed+ mitochondria (right). Following photobleaching, cells are imaged over time to quantify the amount of cell division. (h) Quantification of cell division after photobleaching. Each data point is the average within a field of view (N=13 experiments), with control (cyto) and experimental (mito) data shown as paired dots per experiment. Scale bars are 10 µm. Wilcoxon matched-pairs signed rank test (d, h), two-way ANOVA (f), *p<0.05; ****p<0.0001.
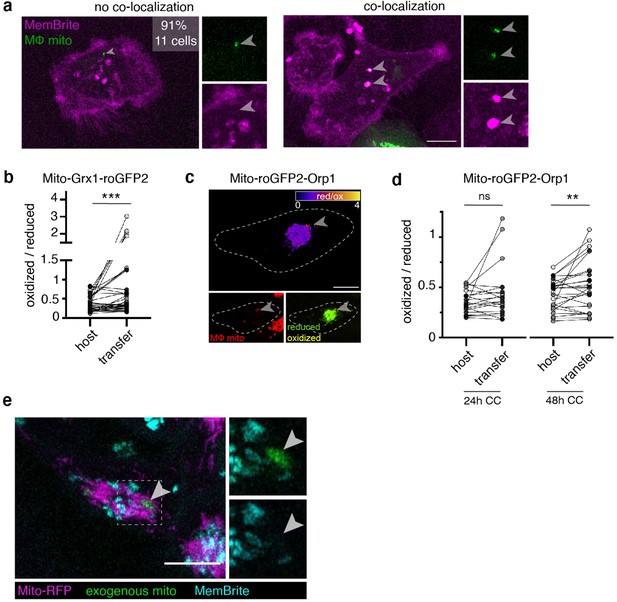
Transferred mitochondria accumulate reactive oxygen species, and internalized exogenous mitochondria are not encapsulated in a membrane compartment.
(a) Recipient 231 cells were stained with a dye, MemBrite, that marks both the plasma and vesicular membranes. 91% of transferred mitochondria (green, arrowheads) did not co-localize with MemBrite signal (magenta, left panel) whereas 9% did (right panel) (N=11 cells, 1 donor). (b) Ratiometric measurements of the mito-Grx1-roGFP2 sensor per 231 cell (paired dots) at a region of interest (ROI) containing the recipient host mitochondrial network (host) or a transferred mitochondria (transfer). Cells were co-cultured with macrophages for 48 hr (N=37 cells, 3 donors), individual donors are indicated as shades of gray. (c) Ratiometric quantification of mito-roGFP2-Orp1 biosensor mapped onto recipient 231 cell in the fire LUT (top). Bottom left panel shows macrophage mitochondria (bottom left, red, arrowhead) and bottom right shows mito-roGFP2-Orp1 (green and yellow). (d) Ratiometric measurements of the mito-roGFP2-Orp1 sensor per 231 cell (paired dots) at a ROI containing the host mitochondrial network (host) or a transferred mitochondria (transfer). Cells were co-cultured with macrophages for 24 hours (N=21 cells, 3 donors, left) or 48 hr (N=26 cells, 3 donors, right).( e), Mitochondria were isolated from mito-mEm expressing THP-1 cells. The purified mitochondrial preparations (green) were perfused onto MDA-MB-231 cells expressing mito-RFP (magenta). 24 hr after mitochondrial uptake, MDA-MB-231 cells were stained with MemBrite (cyan) to visualize membranes. Longer incubations with MemBrite stain allow for visualization of internal membrane structures (intracellular cyan-positive structures in the image), and internalized exogenous mitochondria did not colocalize with MemBrite staining (arrowheads). Individual donors are indicated as shades of gray. All scale bars are 10 μm. Wilcoxon matched-pairs signed rank test (b, d), **p<0.01; ***p<0.0001.
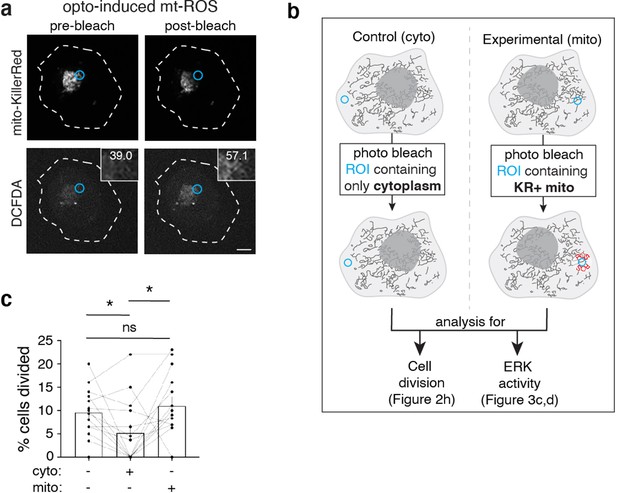
Inducing reactive oxygen species results in cancer cell proliferation.
(a) Photobleaching a region of interest (ROI; blue) containing KillerRed + mitochondria (top panels) generate an increase in ROS levels (DCFDA, ROS indicator, bottom panels). Mean fluorescent intensity (MFI) of DCFDA list in the inset. (b) Schematic of optogenetic experiments to generate data in Figure 2g and h and Figure 3c and d. Cells expressing mito-KillerRed are photobleached in a specific ROI containing either cytoplasm only (left) or mito-KillerRed+ mitochondria (right). Following photobleaching, cells are imaged over time to quantify the amount of cell division (Figure 2g and h) or ERK activity (Figure 3c and d). (c) Data from Figure 2h plotted next to an additional control. Quantification of percent cells divided after no stimulation (left column), photobleaching an ROI containing cytoplasm (cyto, middle column), or an ROI containing mito-KillerRed+ mitochondria (mito, right column). Each data point is the average within a field of view per condition (N=13 experiments). Error bars represent SEM and scale bars are 10 μm. Wilcoxon matched-pairs signed rank test, *p<0.05.
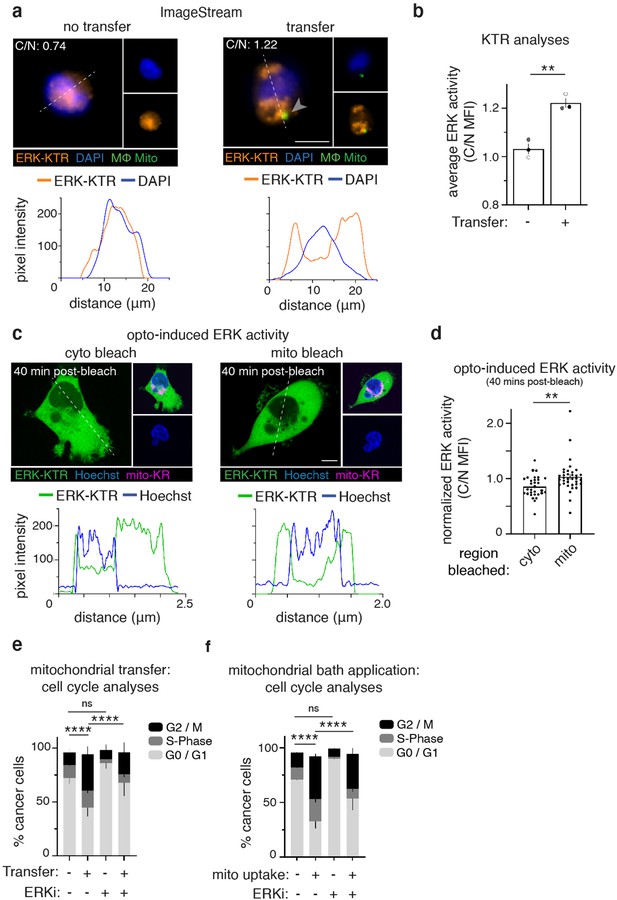
Recipient cancer cells exhibit ERK-dependent proliferation.
(a) ImageStream was used to measure the MFI of an ERK-Kinase Translocation Reporter (ERK-KTR, orange) in the nucleus (DAPI, blue) or cytoplasm of co-cultured 231 cells that did (right) or did not (left) receive mitochondria (green, arrowhead). Below: representative line scans (white dotted lines) of ERK-KTR (orange) and DAPI (blue). (b) Average ERK activity from data displayed in (d) (cytoplasm/nucleus (C/N) mean fluorescence intensity (MFI); N=3 donors indicated as shades of gray). (c) Confocal images of 231 cells expressing ERK-KTR (green) and Mito-KillerRed (magenta) with Hoechst 33342 (blue), after control cytoplasmic bleach (cyto, left) or mito-KillerRed+ bleach (mito, right). Below: representative line scans (white dotted lines) of ERK-KTR (green) and Hoechst (blue). (d) Quantification of ERK-KTR translocation 40 min post-bleach (cyto vs. mito), normalized to time 0. Each dot represents a measurement from a single cell. (e) Analysis of proliferative capacity by quantifying Ki-67 and DNA levels of co-cultured 231 cells treated with vehicle or ERK inhibitor (ERKi) with or without transfer or (f), mitochondrial internalization after mitochondrial bath application (N=3 donors; statistics for G2/M only). Error bars represent SEM and scale bars are 10 µm., Welch’s t-test (b), Mann-Whitney (d), two-way ANOVA (e–f), *p<0.05; **p<0.01; ****p<0.0001.
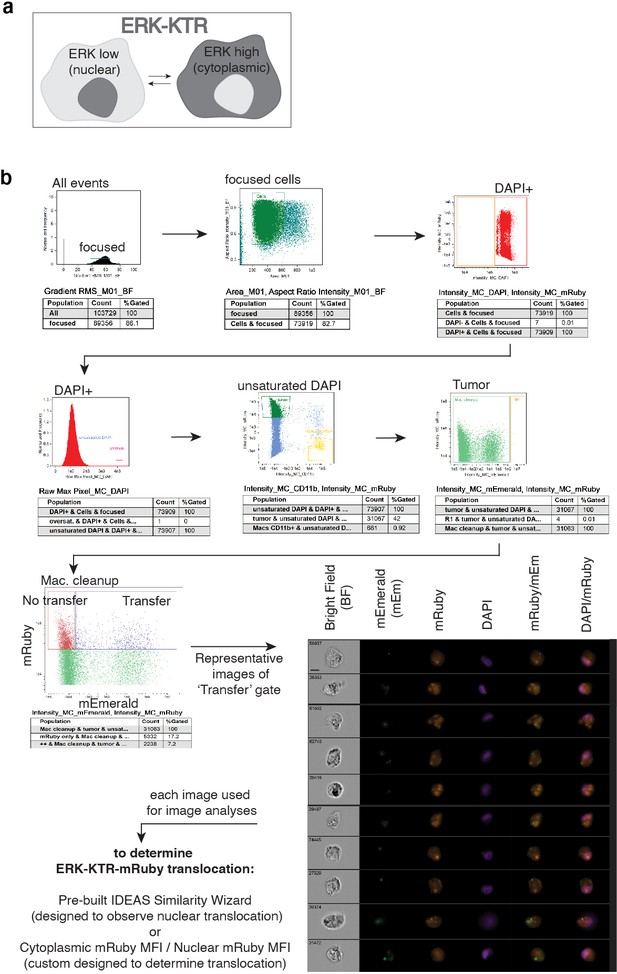
Amnis ImageStream pipeline for ERK-KTR quantification.
(a) Schematic of ERK-Kinase Translocation Reporter (KTR). When ERK activity is low, the fluorescent protein of the KTR resides primarily in the nucleus (left, dark gray). When ERK activity is high, the fluorescent protein translocates to the cytoplasm (right). (b) Workflow of Amnis ImageStream analyses. Mito-mEm macrophages and ERK-KTR-mRuby 231 cells were co-cultured for 24 hr and then analyzed on the ImageStream. Single cells in focus are selected and populations of interest can be isolated for further analyses. Representative images of recipient cells within our ‘transfer’ gate are displayed in all channels acquired (bottom right images). All populations of interest are then put though two independent readouts of KTR translocation: the pre-built IDEAS Similarity Wizard and/or the custom-built cytoplasmic:nuclear (cyto:nuc) mRuby mean fluorescent intensity masking algorithm. Scale bar is 10µm.
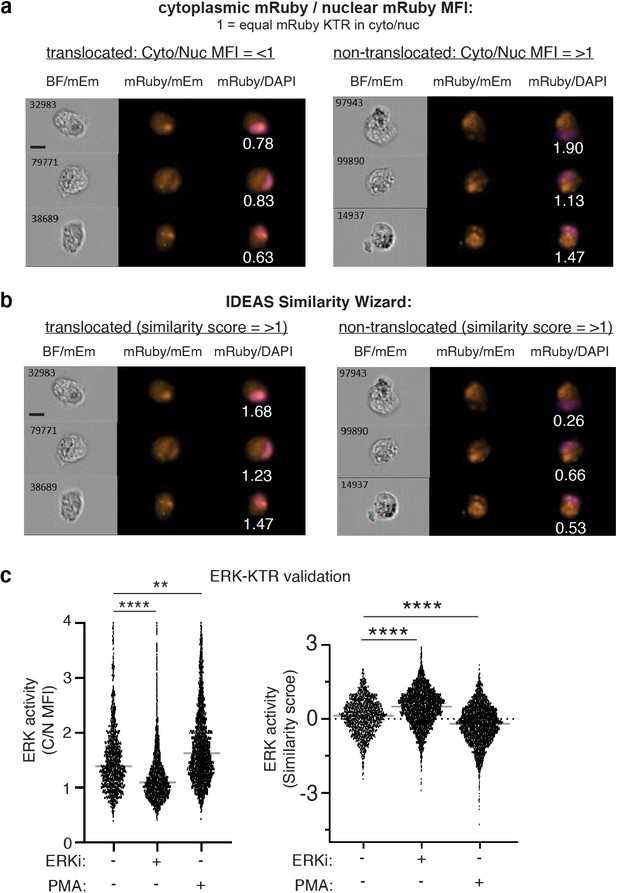
ERK-KTR analysis and validation using the Amnis ImageStream pipeline.
(a) Representative ImageStream images using the mRuby mean fluorescence intensity (MFI) masking algorithm to quantify the amount of cytoplasmic cyto (cyto, C) and nuclear (nuc, N) ERK-KTR-mRuby. Representative images of translocated (left) and non-translocated (right) ERK-KTR images are displayed with the ratiometric cyto/nuc values listed below the corresponding images. (b) Representative ImageStream images using the IDEAS Similarity Wizard to quantify how similar ERK-KTR fluorescent signal is to the nuclear signal (DAPI). Representative images are displayed showing examples of translocated (left) and non-translocated (right) images with the ratiometric values listed below the corresponding images. (c) Quantification of ERK-KTR translocation in 231 monocultures using ImageStream after treatment with 1 μM of an ERK inhibitor (ERKi), SCH772984, or an ERK stimulator, Phorbol 12-myristate 12-acetate (PMA). Cyto/Nuc MFI of ERK-KTR (left) and similarity scores (right) for the same data set are displayed (N=1 donor). Scale bars are 10µm. One-way ANOVA, **p<0.01; ****p<0.0001.
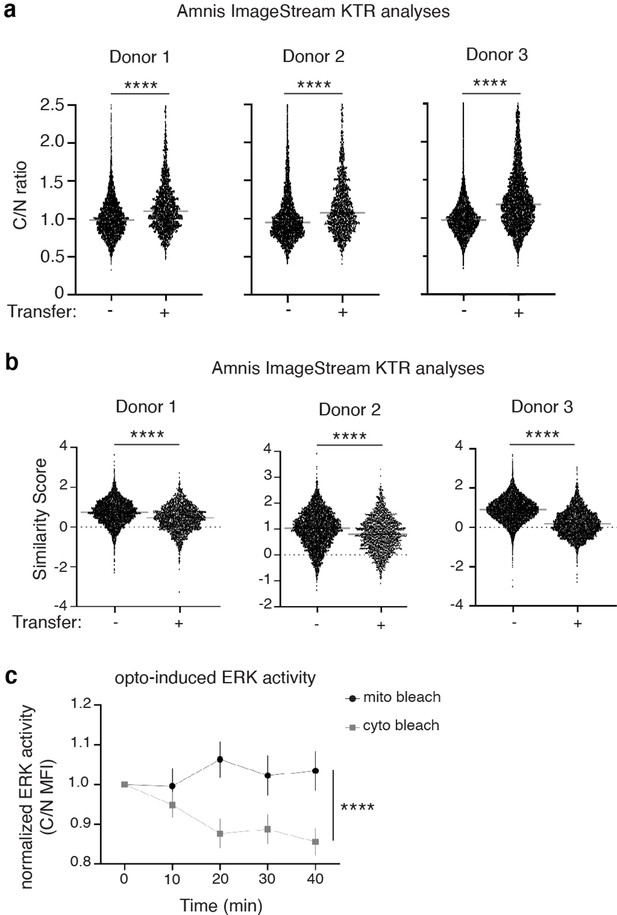
Quantification of ERK activity in recipient 231 cells or upon ROS induction.
(a) Cytoplasmic/Nuclear mean fluorescence intensity (C/N MFI) ratios of the ERK-KTR in co-cultured 231 cells are displayed for three different macrophage donors (N=3 experiments). Each data set shows an increase in C/N MFI ratios in recipient cells (left column on each plot) indicating higher ERK activity compared to their non-recipient co-cultured counterparts. (b) The same three experimental samples from the analyses in (a) were analyzed using the IDEAS Similarity Wizard. This analysis indicates cells that receive transfer (right column on each plot) have a lower similarity score, indicating higher ERK activity compared to cells that did not receive transfer (N=3 experiments). (c) Quantification over time of ERK-KTR translocation after mito bleach (black circles) compared to control cyto bleach (gray squares). Welch’s t-test (a–b), Two-way ANOVA at time point 40 min (c), ****p<0.0001.
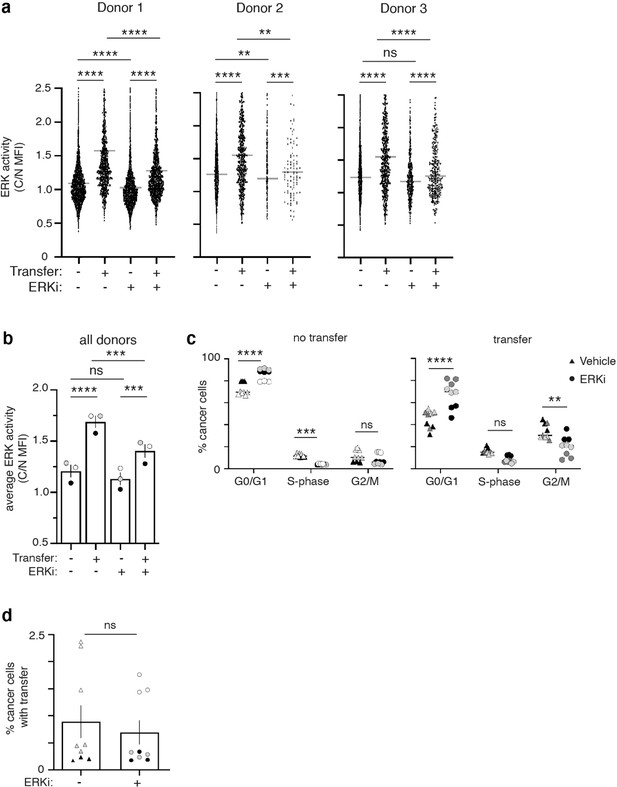
ERK inhibition reduces proliferation in cancer cells with macrophage mitochondria.
(a) Co-cultured 231 cells treated with 1 μM ERKi or vehicle at time of plating were analyzed for ERK-KTR translocation. Data are displayed from 3 different macrophage donors (N=3 experiments). Treatment with ERKi significantly reduced the amount of ERK activity in cells that did or did not receive transfer. (b) Average ERK activity from all experiments shown in (a) is displayed (N=3 donors). (c) Co-cultured macrophages and 231 cells were treated with 1 μM ERKi or vehicle at time of plating. Flow cytometry was used to determine how treatment influenced the cell cycle. Plots compare 231 cells that did (right) and did not (left) receive macrophage mitochondria in each stage of the cell cycle treated with vehicle (triangles) or ERKi (circles) (N=3 donors in technical triplicate). (d) Co-cultured 231 cells were treated with ERKi or vehicle at time of plating and the rate of mitochondrial transfer was determined with flow cytometry (N=3 donors in technical triplicate). For all panels, individual donors are indicated as shades of gray and error bars represent SEM. One-way ANOVA (a), two-way ANOVA (b–d), *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
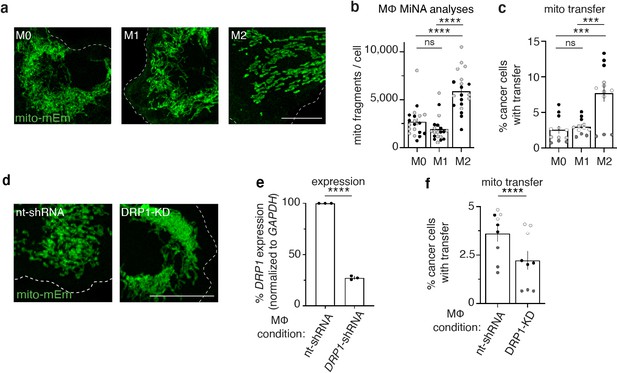
M2-like macrophages exhibit increased mitochondrial fragmentation and increased mitochondrial transfer to cancer cells.
(a) Representative images of mito-mEm+ macrophages that were non-stimulated (M0, left) or activated to become M1-like (middle) or M2-like (right). (b) Mitochondrial network analyses (MiNA) were used to determine number of mitochondrial fragments per cell (N=2 donors). (c) Macrophages were co-cultured with mito-RFP 231 cells for 24 hr and mitochondrial transfer was quantified with flow cytometry (N=4 donors). (d) Representative images of mito-mEm (green) macrophages in macrophages with control nt-shRNA KD and DRP1 KD. (e) q-RT-PCR of DRP1 knockdown (DRP1-KD) macrophages (N=3 donors). (f) Rates of mitochondrial transfer with control and DRP1-knockdown macrophages (N=3 donors). For all panels, individual donors are indicated as shades of gray with each cell as a data point, error bars represent SEM and scale bars are 10 µm. Two-way ANOVA (b, c), unpaired t-test (e, f), ***p<0.001; ****p<0.0001.
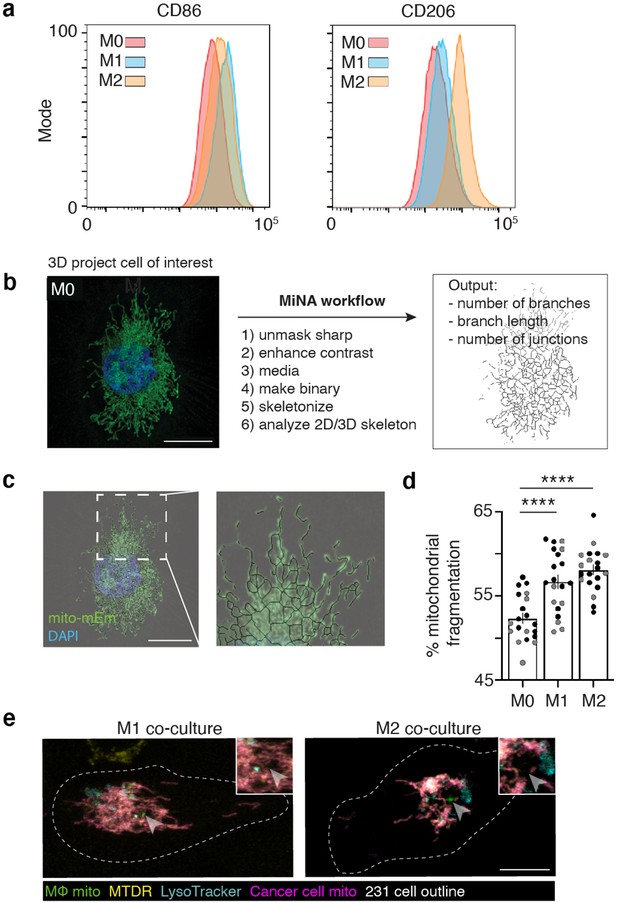
M2-like macrophages exhibit increased mitochondrial transfer to cancer cells.
(a) Macrophages were activated with IFN-γ (M1 activation; left) or IL-4/IL-13 (M2 activation; right) for 48 hours and flow cytometry was used to determine expression of canonical M1 (CD86, left) and M2 (CD206, right) markers. Representative histograms shown. (b) Mitochondrial Network Analyses (MiNA) workflow with a representative input confocal image (left) and example of skeletonized mitochondrial network (right) to quantify number of branches per individual mitochondria, branch length, and number of junctions per individual mitochondrion. (c) Representative confocal image with a post-processed skeletonized version of network overlaid. (d) Percent mitochondrial fragmentation in M0, M1-like, and M2-like macrophages (N=2 donors). Two-way ANOVA, ****p<0.0001. (e) Representative confocal images of mito-RFP 231 cells co-cultured with mito-mEm M1-like (left) or M2-like (right) macrophages and stained with MitoTracker Deep Red (MTDR, yellow) and LysoTracker (LT, Teal). 100% of transferred mitochondria (green, arrowhead) from M1-like and M2-like macrophages are MTDR-negative. 63.6% (from M1-like) and 48% (from M2-like) of transferred mitochondria do not co-localize with LT (For M1-like MTDR and LT staining, N=15 cells, 2 donors; for M2-like MTDR staining, N=24 cells, 2 donors; for M2 LT staining, N=17 cells, 2 donors). For all panels, individual donors are indicated as shades of gray, error bars represent SEM and scale bars are 10 μm.
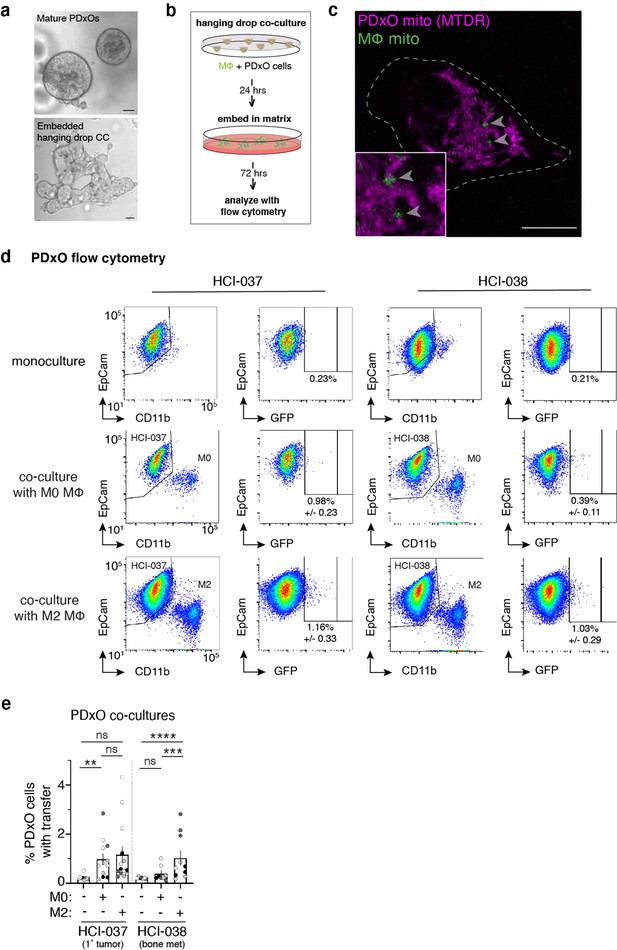
Macrophages transfer mitochondria to breast cancer patient-derived cells.
(a) Representative images of the HCI-037 patient-derived xenograft organoid (PDxO) line in culture (top) or an embedded hanging drop co-culture with macrophages (bottom). (b) Schematic of experimental setup of PDxOs (gray)/mito-mEm macrophages (green) co-cultures. Co-cultures are plated in suspended drops of media (hanging drops) to allow for the formation of cell aggregates without adherence to a substrate. After 24 hr, the co-cultured cells are embedded into a matrix (Matrigel) and cultured for 72 hr before analysis with flow cytometry. (c) Representative image of a FACS-isolated PDxO cell containing macrophage mitochondria (green, arrowhead) that are MTDR-negative. (d) Top row: Representative flow cytometry plot of PDxO monocultured cells used as the experimental control when quantifying mitochondrial transfer. Middle row: PDxO cells co-cultured with mito-mEm-expressing M0 macrophages. Bottom row: PDxO cells co-cultured with mito-mEM expressing M2-like macrophages. PDxO lines used for co-culture are indicated at the top of the corresponding panels. Scale bars are 10 μm. (e), Rate of mitochondrial transfer to HCI-037 (left 3 columns) or HCI-038 (right 3 columns) PDxO cells from M0 or M2 macrophages (each dot is one replicate, N=4 donors). Two-way ANOVA, **p<0.01; ***p<0.001; ****p<0.0001.
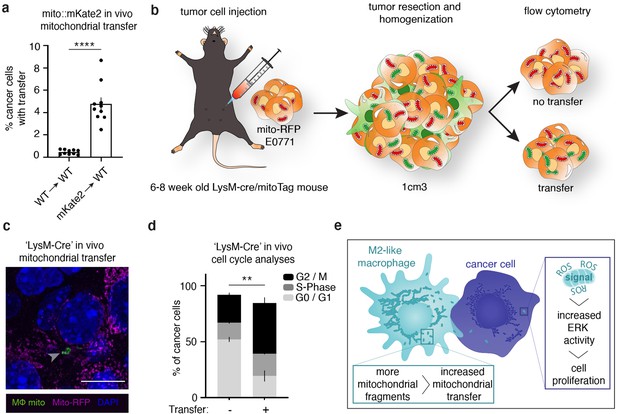
Macrophage mitochondrial transfer promotes tumor cell proliferation in vivo.
(a) Quantification of E0771 mammary adenocarcinoma cells from in vivo tumors with mKate2+ mitochondria in bone marrow reconstitution experiments versus control mice. N=10 mice per condition. (b) Schematic representation of a second mouse model to quantify proliferation in cancer cells with macrophage mitochondria in vivo. Myeloid lineages were specifically labeled with mito-GFP by crossing a Loxp-Stop-Loxp-MitoTag-GFP mouse to a LysM-Cre mouse. E0771 cells expressing mito-RFP were injected into the mammary fat pad of mice with MitoTag-GFP expression in myeloid cells, and tumors were isolated and analyzed for direct observation of transfer through fluorescent microscopy (c) and Ki67/DNA to quantify proliferative index (d). (c) Representative immunofluorescence image of E0771 tumor cell expressing mito-RFP (magenta) containing GFP+ macrophage mitochondria (arrowheads) from mice in which GFP+ mitochondria are restricted to the myeloid lineage (‘LysM-Cre’). (d) Cell cycle analysis of E0771 in vivo tumor cells with and without GFP+ macrophage mitochondria in ‘LysM-Cre’ model in which GFP+ mitochondria are restricted to the myeloid lineage. N=3 mice. (e) Working model for macrophage mitochondrial transfer to breast cancer cells. For all panels, individual donors are indicated as shades of gray with each cell as a data point, error bars represent SEM and scale bars are 10 µm. Welch’s t- test (a), two-way ANOVA (d), **p<0.01; ****p<0.0001.
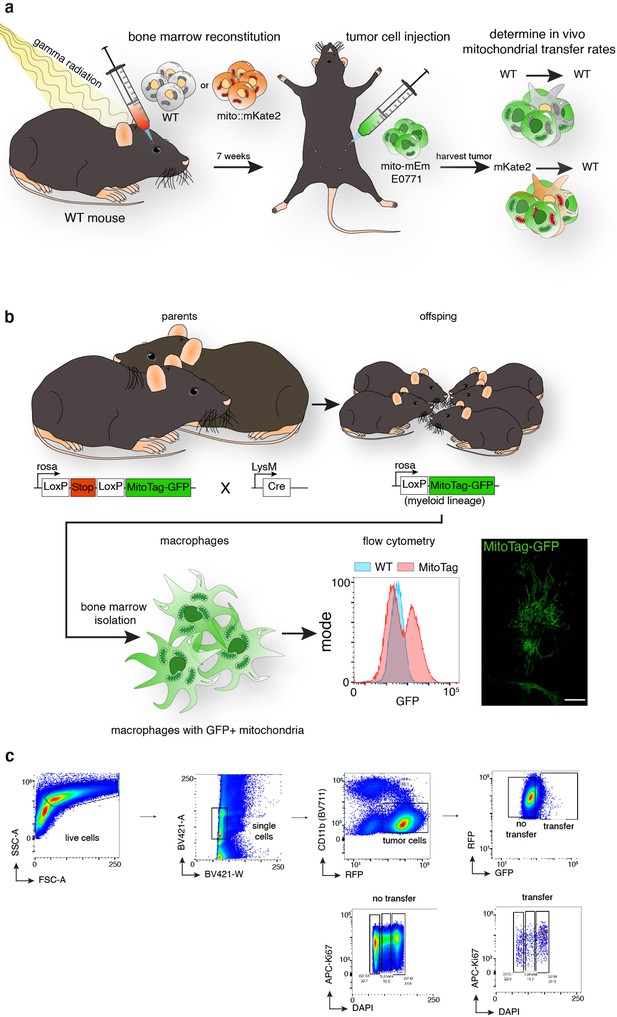
Murine mammary adenocarcinoma cells with macrophage mitochondria exhibit increased cell proliferation in vivo.
(a) Schematic representation of mouse model to quantify macrophage mitochondrial transfer. Bone marrow from either mito::mKate2 or wildtype mice are transplanted into gamma-irradiated mouse hosts. Once grafted, E0771 cells expressing mito-mEm are injected into the mammary fat pad of these mice, and tumors were isolated and analyzed for mitochondrial transfer by flow cytometry. (b) Schematic representation of a second mouse model to quantify proliferation in cancer cells with macrophage mitochondria in vivo. Myeloid lineages were specifically labeled with mito-GFP by crossing a Loxp-Stop-Loxp-MitoTag-GFP mouse to a LysM-Cre mouse. (c) Gating strategy for Ki67/DNA analysis for E0771 tumors.
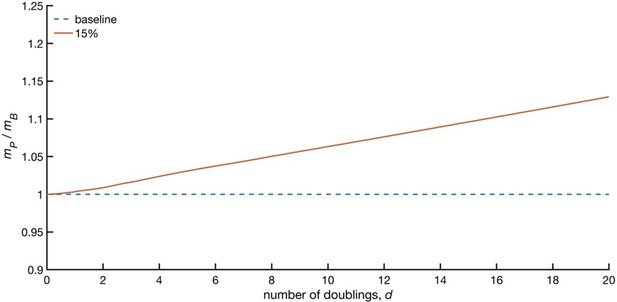
Predicted increase in population size due to transferred mitochondria as a function of number of population doublings.
Model results are presented as the ratio of mP, the mass of the population receiving macrophage mitochondria to mB, the mass of the population with baseline growth rate. The initial fraction of cells with transferred mitochondria and the fraction of the population receiving mitochondria, f, over any cell division cycle is assumed to be 5%. 50% of cells lose mitochondria at division. The population was initially seeded with 100 cells and an even distribution of mass between m0 and 2 m0, the baseline initial mass.
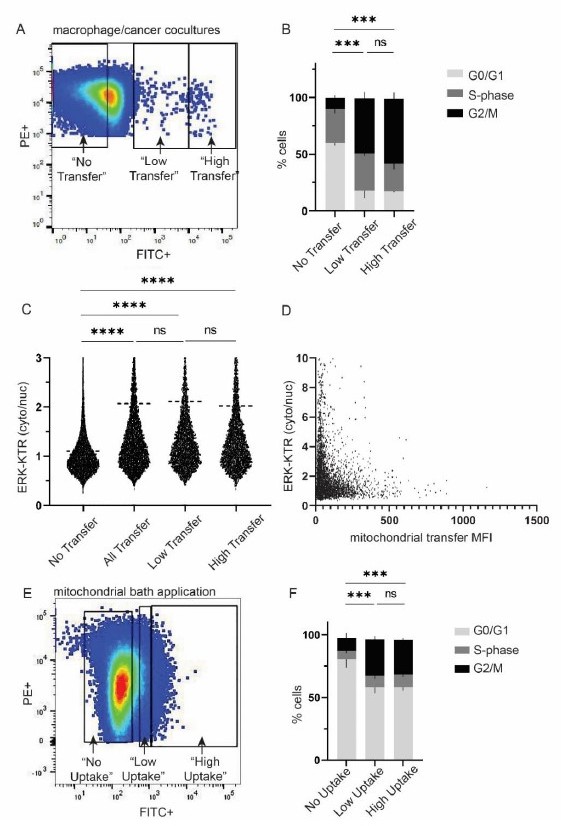
A.
Representative gating strategy for “low” vs “high” macrophage mitochondrial transfer in macrophage/cancer cell cocultures. B. Cell cycle analysis for populations in (A). 2-way ANOVA, n=2 biological replicates, p<0.001. C. ERK-KTR analysis with imagestream, n=3 biological replicates, 1-way ANOVA, p<0.0001. D. Scatterplot of ERK-KTR and mitochondrial transfer mean fluorescence intensity (MFI). Each dot is a cell. n=7,966 cells. Correlation R2 value: 3.261e-005. E. Representative gating strategy for “low” vs “high” macrophage mitochondrial uptake by cancer cells. F. Cell cycle analysis for populations in (E). 2-way ANOVA, n=3 biological replicates, p<0.001.
Videos
Macrophage mitochondria are long-lived and remain distinct in recipient cancer cells.
Video depicting a recipient mito-RFP expressing 231 cell (magenta) that contains mito-mEm macrophage mitochondria (green in magenta cell, center of frame). 231 cells were co-cultured with macrophages for 7 hr prior to the start of imaging for a duration ~15 hr with a time interval of 5 min. Maximum intensity projections of images are displayed at 12 frames per second, timestamp in upper left corner in hours (h), and scale bar is 10 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (M. musculus) | wildtype C57BL/6 J | The Jackson Laboratory | Stock #000664 | |
| strain, strain background (M. musculus) | mito:mKate2 mouse Tg(CAG-mKate2)1Poche/J | The Jackson Laboratory | Stock 032188 | |
| strain, strain background (M. musculus) | LysM-Cre mouse B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Stock 004781 | |
| strain, strain background (M. musculus) | Lox-stop-lox-MitoTag mouse B6N.Cg-Gt(ROSA)26Sortm1(CAG-EGFP*)Thm/J | The Jackson Laboratory | Stock 032675 | |
| cell line (Homo-sapiens) | MDA-MB-231 | American Type Culture Collection | HTB-26 | |
| cell line (Homo-sapiens) | MDA-MB-468 | American Type Culture Collection | HTB-132 | |
| cell line (Homo-sapiens) | A375 | American Type Culture Collection | CRL-1619 | |
| cell line (Homo-sapiens) | THP-1 | American Type Culture Collection | TIB-202 | |
| cell line (Homo-sapiens) | MCF10a | American Type Culture Collection | CRL-10317 | |
| cell line (M. musculus) | E0771 | American Type Culture Collection | CRL-3461 | |
| transfected construct (Homo-sapien) | pLPCX mito-Grx1-roGFP2 | Addgene | 64977 | |
| transfected construct (S. cerevisiae) | pLPCX mito-roGFP2-Orp1 | Addgene | 64992 | |
| antibody | BV711-CD11b (mouse anti-human, monoclonal) | Biolegend | 301344 | 1:20-1:40 Used for flow cytometry |
| antibody | PE anti-human CD326 (EpCAM) Antibody (mouse andti-human, monoclonal) | Biolegend | 369806 | 1:40 Used for flow cytometry |
| antibody | APC-Ki67 (mouse anti-human, Monoclonal) | ThermoFisher | 17-5699-42 | 1:20 – 1:40 Used for flow cytometry |
| antibody | anti-GFP (Chicken, polyclonal) | Abcam | AB13970, | 1:500 |
| antibody | anti-RFP (rabbit, polyclonal) | Abcam | AB62341 | 1:1000 |
| antibody | Alexa Fluor 488 AffiniPure (Goat anti-Chicken, polyclonal) | Jackson ImmunoResearch | 103-545-155 | 1:500 |
| antibody | IgG (H+L) (Cross-Adsorbed Goat anti-Rabbit Alexa Fluor 555, polyclonal) | Invitrogen | A21428 | 1:500 |
| recombinant DNA reagent | Mito-mEm: pLKO.1 mito-mEmerald | This paper | Addgene, 174548 | Lentiviral construct to transfect and express fluorescently-tagged mitochondria |
| recombinant DNA reagent | Mito-RFP: pLKO.1 mito-TagRFP-T | This paper | Addgene, 174543 | Lentiviral construct to transfect and express fluorescently-tagged mitochondria |
| recombinant DNA reagent | mEmerald-TOMM20: pLKO.1 mEmerald-TOMM20-N-10 | This paper | Addgene, 54282 | Lentiviral construct to transfect and express fluorescently-tagged mitochondria |
| recombinant DNA reagent | mCherry-TOMM20: pLKO.1 mCherry-TOMM20-N-10 | This paper | Addgene, 55146 | Lentiviral construct to transfect and express fluorescently-tagged mitochondria |
| recombinant DNA reagent | Mito-KR: pLKO.1 3xHA-KillerRed-OMP25 | This paper | Addgene, 174544 | Lentiviral construct to transfect and express mitochondrially-localized KillerRed |
| recombinant DNA reagent | ERK-KTR-mRuby: pLentiPGK Blast DEST ERKKTRmRuby2 | Addgene | 90231 | Lentiviral construct to transfect and express ERK Kinase Translocation reporter |
| recombinant DNA reagent | ERK-KTR-Clover: pLentiPGK Puro DEST ERKKTRClover | Addgene | 90227 | Lentiviral construct to transfect and express ERK Kinase Translocation reporter |
| recombinant DNA reagent | Non-target-shRNA | Sigma | SHC002 | Lentiviral construct to transfect and express non-target shRNA |
| recombinant DNA reagent | DRP1-KD: DRP1-shRNA | Sigma | TRCN0000001097 | Lentiviral construct to transfect and knock down gene target HGNC ID 2973 |
| sequence-based reagent | Primer: DRP1-F | This paper | AGAAAATGGGGTGGAAGCAGA | |
| sequence-based reagent | Primer: DRP1-R | This paper | AAGTGCCTCTGATGTTGCCA | |
| sequence-based reagent | Primer: GAPDH-F | This paper | AGCCACATCGCTCAGACA | |
| sequence-based reagent | Primer: GAPDH-R | This paper | ACATGTAAACCATGTAGTTGAGGT | |
| peptide, recombinant protein | GM-CSF | Peprotech | 300–03 | 20 ng/mL |
| peptide, recombinant protein | IFN-γ | Peprotech | 3000–02 | 20 ng/mL |
| peptide, recombinant protein | IL-4 | Peprotech | 200–04 | 20 ng/mL |
| peptide, recombinant protein | IL-13 | Peprotech | 200–13 | 20 ng/mL |
| commercial assay or kit | eBioscience Foxp3/Transcription Factor Staining Buffer Set | ThermoFisher | 00-5523-00 | |
| commercial assay or kit | Polyplus-transfection jetPRIME DNA/siRNA transfection kit | Genesee Scientific | 55–131 | Used to transfect pLPCX mito-Grx1-roGFP2 and pLPCX mito-roGFP2-Orp1 probes |
| commercial assay or kit | MitoTracker Deep Red | ThermoFisher | M22426 | Used at 25 nM |
| commercial assay or kit | TMRM: Tetramethylrhodamine, Methyl Ester, Perchlorate | ThermoFisher | T668 | Used at 100 nM |
| commercial assay or kit | LysoTracker Blue | ThermoFisher | L7525 | Used at 75 nM |
| commercial assay or kit | MemBrite 640/660 | Biotium | 30097 | Used at 1:1000 |
| commercial assay or kit | DCFDA: Carboxy-H2DCFDA | ThermoFisher | C400 | Used at 5μM |
| chemical compound, drug | ERKi: SCH772984 | SelleckChem | 7101 | Used at 1 µM |
| chemical compound, drug | PMA: Phorbol 12-myristate 13-acetate | SelleckChem | S7791 | Used at 100 nM (cancer cell treatment) and 162 nM (THP1 differentiation) |
| chemical compound, drug | MitoTEMPO | Cayman Chemical | 16621 | Used at 100 µM |
| software, algorithm | QPI analyses | This Paper | https://github.com/Zangle-Lab/Macrophage_tumor_mito_transfer | |
| software, algorithm | Single-cell RNA-sequencing | This paper | GEO accession number: GSE181410 (RRID:SCR_002630 (version number 1)) https://github.com/rohjohnson-lab/kidwell_casalini_2021 |