Structural screens identify candidate human homologs of insect chemoreceptors and cryptic Drosophila gustatory receptor-like proteins
Figures
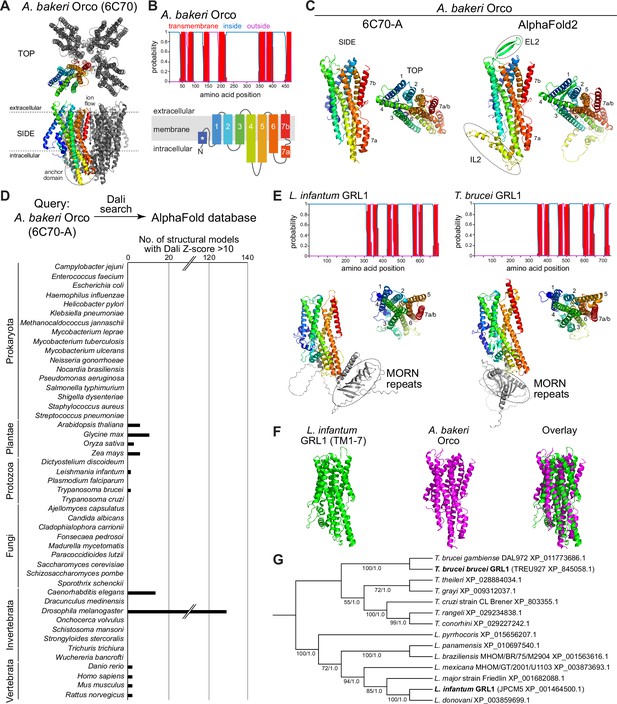
Structure-based screening for seven transmembrane domain ion channel (7TMIC) homologs.
(A) Top view of a cryo-electronic microscopic (cryo-EM) structure of the homotetramer of Or co-receptor (Orco) from A. bakeri (derived from PDB 6C70; Butterwick et al., 2018), in which one subunit has a spectrum coloration (N-terminus [blue] to C-terminus [red]). The ion channel pore is formed at the interface of the four subunits. A side view is shown below. The anchor domain, comprising the cytoplasmic projections of TM4-6 and TM7a, forms most of the inter-subunit interactions in odorant receptors (Ors) (Butterwick et al., 2018; Del Mármol et al., 2021). (B) Top: output of transmembrane topology predictions of DeepTMHMM (Hallgren et al., 2022) for A. bakeri Orco. Bottom: schematic of the membrane topology of an Orco monomer, with the same spectrum coloration as in (A), reproduced from Figure 1a from Benton et al., 2020. Note that the seventh predicted helical region is divided into two in the cryo-EM structure: TM7a (located in the cytosol) and TM7b (located in the membrane). (C) Comparisons of side and top views of the cryo-EM structure of an A. bakeri Orco subunit (6C70-A) (left) and an AlphaFold2 protein structure prediction of A. bakeri Orco. Helical regions are numbered in the top views. Note the model contains the extracellular loop 2 (EL2) and intracellular loop 2 (IL2) regions that were not able to be accurately visualized in the cryo-EM structure (Butterwick et al., 2018). Quantitative comparisons of structures are provided in Table 1. (D) Summary of the results of the screen for Orco/Or-like protein folds in the AlphaFold Protein Structure Database for the indicated species using Dali (Holm, 2022). The threshold of Dali Z-score >10 was informed by inspection of the results of the screen (see Results). Raw outputs of the screen are provided in Source data 2. (E) Top: transmembrane topology predictions of the single screen hits from the Trypanosoma species Leishmania infantum and Trypanosoma brucei brucei. Bottom: AlphaFold2 structural models of these proteins, displayed as in (C). The long N-terminal region contains tandem Membrane Occupation and Recognition Nexus (MORN) repeats and sequence of unknown structure (gray); these are masked in the top view of the models. (F) Visual comparison of the L. infantum GRL1 AlphaFold2 model (the N-terminal region is masked) with the A. bakeri Orco structure, aligned with Coot (Emsley et al., 2010). Quantitative comparisons of structures are provided in Table 1. (G) Consensus phylogeny of putative trypanosome homologs. The primary sequence database was assembled using L. infantum GRL1 (XP_001464500.1) and T. brucei brucei GRL1 (XP_845058.1) as query sequences (highlighted in bold). Branch support values refer to maximum likelihood UFboot/Bayesian posterior probabilities. Note that although the Trypanosoma cruzi homolog (XP_803355.1) was not identified in the original Dali screen, visual inspection of the corresponding AlphaFold2 model (A0A2V2WL40) revealed the same global fold.
-
Figure 1—source data 1
FASTA file containing the amino acid sequences for validated trypanosome GRLs used in phylogenetic analyses.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig1-data1-v2.zip
-
Figure 1—source data 2
FASTA file containing the multiple sequence alignment of trypanosome GRLs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig1-data2-v2.zip
-
Figure 1—source data 3
Newick tree file containing the maximum likelihood phylogeny of trypanosome GRLs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig1-data3-v2.zip
-
Figure 1—source data 4
NEXUS tree file containing the Bayesian phylogeny of trypanosome GRLs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig1-data4-v2.zip
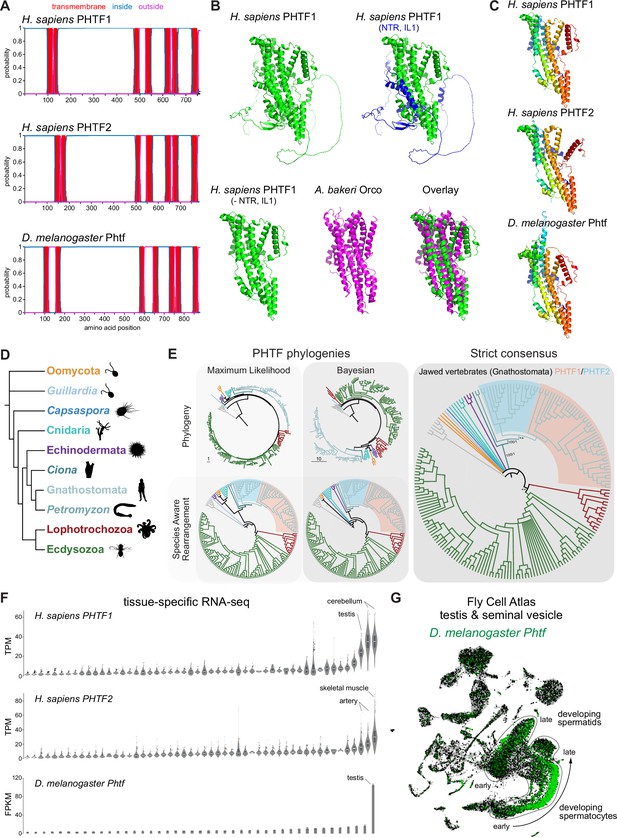
PHTF proteins are candidate vertebrate seven transmembrane domain ion channels (7TMICs).
(A) DeepTMHMM-predicted transmembrane topology of PHTF proteins. (B) Top: AlphaFold2 predicted structure of H. sapiens PHTF1; in the image on the right the long N-terminal region (NTR) and intracellular loop 1 (IL1) are highlighted in blue; these sequences contain a few predicted helical regions but are of largely unknown structure. Bottom: visual comparison of the H. sapiens PHTF1 AlphaFold2 structure (in which the NTR and IL1 are masked) with the A. bakeri Or co-receptor (Orco) structure. (C) AlphaFold2 structures of PHTF proteins in which the NTR and IL1 are masked. Quantitative comparisons of these structures to the cryo-electronic microscopic (cryo-EM) Orco structure are provided in Table 1. (D) Major taxa/species in which a PHTF homolog was identified (see sequence databases in Figure 2—source data 1). Silhouette images in this and other figures are from Phylopic (https://www.phylopic.org/). (E) Phylogenies of a representative set of PHTF sequences. The sequence database was constructed using the D. melanogaster and H. sapiens PHTF query sequences. Top left: maximum likelihood phylogeny (JTT + R10 model) and Bayesian phylogeny. The scale bars represent the average number of substitutions per site. Bottom left: phylogenies where weakly supported branches (<95/0.95) have been rearranged and polytomies resolved in a species tree-aware manner. Right: strict consensus of the species tree-aware phylogenies. There is a single eukaryotic PHTF clade and the PHTF1-2 split occurred in the jawed vertebrate lineage. However, this interpretation relies on the rearrangement of the weakly supported jawless vertebrate PHTF branch. Therefore, an alternative but weakly supported hypothesis is that the duplication occurred in a common vertebrate ancestor and a single PHTF copy was lost in jawless vertebrates. Select branch support values are present on key branches and refer to maximum likelihood UFboot/Bayesian posterior probabilities. Asterisks indicate that branch support was below the threshold for species-aware rearrangement. The fully annotated trees are available in Figure 2—figure supplements 1–3. (F) Summary of tissue-enriched RNA expression of H. sapiens PHTF1 and PHTF2 (data are from the GTex Portal; the fully annotated dataset is provided in Figure 2—figure supplement 4) and D. melanogaster Phtf (data from the Fly Atlas 2.0; the fully annotated dataset is provided in Figure 2—figure supplement 5). (G) Left: Uniform Manifold Approximation and Projection (UMAP) representation of RNA-seq datasets from individual cells of the D. melanogaster testis and seminal vesicle generated as part of the Fly Cell Atlas (10× relaxed dataset) (Li et al., 2022) colored for expression of Phtf. Simplified annotations of cell clusters displaying the highest levels of Phtf expression are adapted from Li et al., 2022; unlabeled clusters represent non-germline cell types of the testis.
-
Figure 2—source data 1
FASTA file containing the amino acid sequences of validated eukaryotic PHTFs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data1-v2.zip
-
Figure 2—source data 2
FASTA file containing the representative amino acid sequences of eukaryotic PHTFs used in phylogenetic analyses.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data2-v2.zip
-
Figure 2—source data 3
FASTA file containing the multiple sequence alignment of eukaryotic PHTFs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data3-v2.zip
-
Figure 2—source data 4
Newick tree file containing the maximum likelihood phylogeny of eukaryotic PHTFs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data4-v2.zip
-
Figure 2—source data 5
NOTUNG tree file containing the species-aware phylogeny of eukaryotic PHTFs, based on the maximum likelihood phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data5-v2.zip
-
Figure 2—source data 6
NEXUS tree file containing the Bayesian phylogeny of eukaryotic PHTFs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data6-v2.zip
-
Figure 2—source data 7
NOTUNG tree file containing the species-aware phylogeny of eukaryotic PHTFs, based on the Bayesian phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data7-v2.zip
-
Figure 2—source data 8
Newick tree file containing the strict consensus of the species-aware phylogenies of eukaryotic PHTFs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig2-data8-v2.zip
Fully annotated phylogenetic trees for PHTF homologs.
Sequences are from the protein sequence database generated using D. melanogaster Phtf and H. sapiens PHTF1/2, and are representatives of clusters of 90% sequence identity. For maximum likelihood, the tree was generated using a JTT + R10 substitution model. Branch support values for maximum likelihood (UFboot) and Bayesian analyses (posterior probability) are shown at the branches. The scale bars represent the average number of substitutions per site.
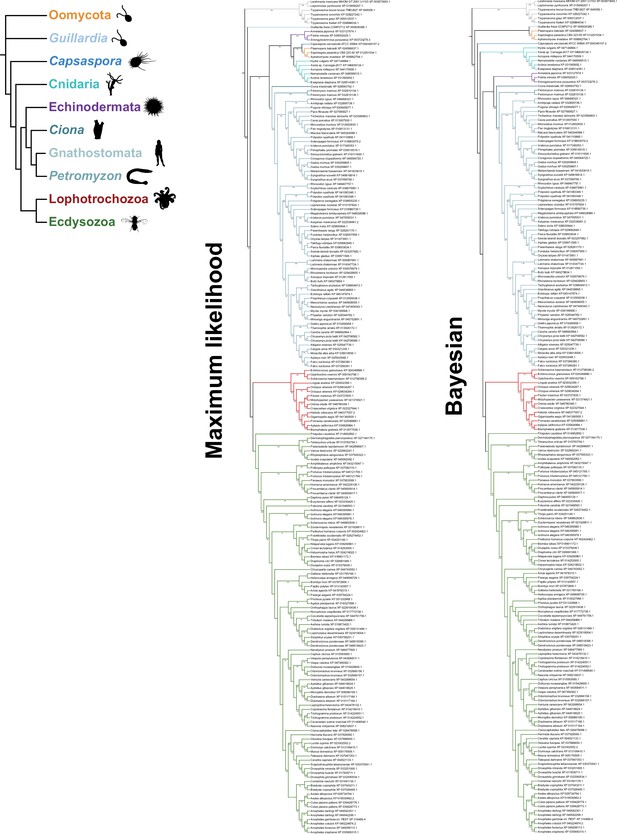
Fully annotated species-aware trees for PHTF homologs.
Trees are based on the maximum likelihood (left) and Bayesian (right) trees. Branches without support values were eligible for rearrangement.
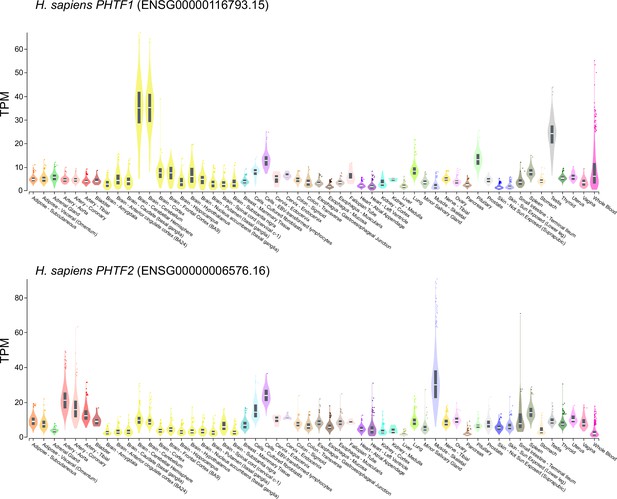
Tissue-specific RNA expression of H. sapiens PHTF1 and PHTF2.
Plot of RNA expression levels (transcripts per million [TPM]) from the indicated tissues is from the GTEx Portal (GTEx Analysis Release V8 [dbGaP Accession phs000424.v8.p2]).
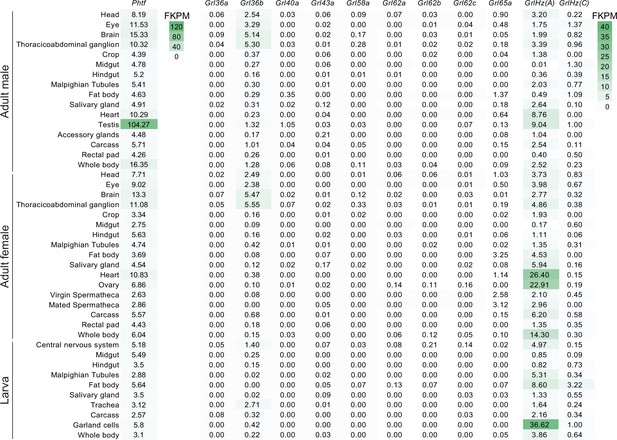
Tissue-specific RNA expression of D. melanogaster Phtf and Grls.
Heatmap plot of the expression of D. melanogaster Phtf and Grls in the indicated tissues/life stages/sexes determined by bulk RNA-seq; fragments per kilobase of exon per million mapped fragments (FPKM) values are shown; data are from the Fly Atlas 2.0 (Krause et al., 2022).
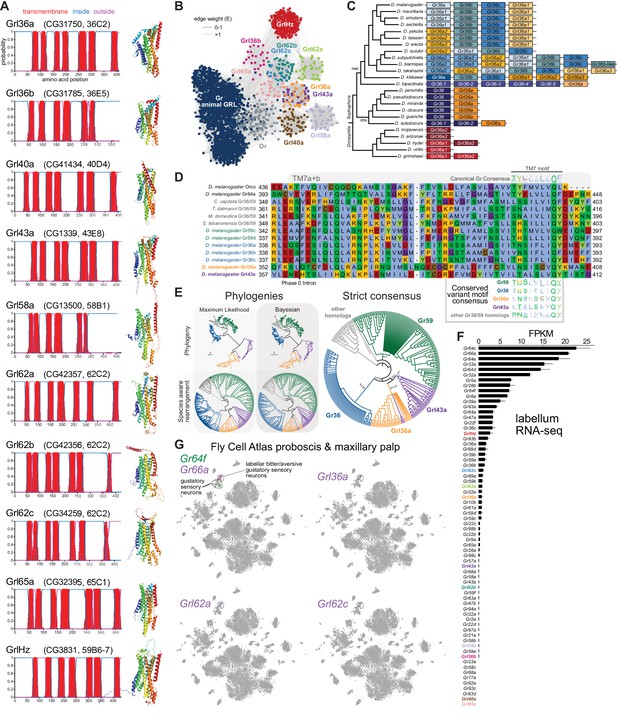
Insect Grls are highly divergent, candidate chemosensory receptors.
(A) Proposed nomenclature of D. melanogaster Grls (the original gene name and cytological location are in parentheses), with corresponding DeepTMHMM-predicted transmembrane topologies and AlphaFold2 structural models. Note that TM7 is not predicted for Grl36b and Grl58a by DeepTMHMM, but is predicted – with the characteristic TM7a/7b split – in the structural model (as well as predicted by Phobius [data not shown]). Quantitative comparisons of these structures to the cryo-electronic microscopic (cryo-EM) Or co-receptor (Orco) structure are provided in Table 1. (B) Sequence similarity network of Grls, Grs, and Ors (including Orco). The network was generated using an all-to-all comparison made by MMSeqs2 as implemented by gs2. The connections represent E-values where the weakest connections (arbitrarily defined as edge weights >1) are colored in lighter gray. Lack of connection between two nodes indicates that those two sequences could not be identified as having any significant sequence similarity under the most sensitive MMSeqs2 settings. Nodes and edges are arranged in a prefuse force-directed layout. The graph splitting tree is visualized in Figure 3—figure supplement 5; however, we do not place high confidence in the phylogenetic accuracy of the tree due to the likely effects of long branch attraction. The evolution of GrlHolozoa (GrlHz) is described in Figure 3—figure supplement 1, with detailed phylogenies in Figure 3—figure supplements 2–4. (C) Schematic of the gene arrangement of Grl36a and Gr36 homologs in drosophilids. Color coding reflects relatedness with respect to major speciation and gene duplication events; colors match the phylogenetic tree branches in Figure 3—figure supplement 6B–C. The Drosophila subgenus entirely lacks Gr36 homologs (see Figure 3—figure supplement 6). (D) Alignment of the C-terminal region of D. melanogaster Orco, Gr64a, select insect Gr36/Gr59 homologs, and D. melanogaster Grl36a and Grl43a, extracted from a larger alignment available in Figure 3—source data 5. The black bar shows the common location of a phase 0 intron, which is presumably homologous in different sequences. The canonical TM7 motif of the Gr family (represented as relative amino acid frequencies extracted from WebLogo) is shown above the sequence, and the variant motifs of different Gr or Grl ortholog groups are shown below. (E) Phylogenies of Gr36, Gr59c/d, Grl36a, Grl43a and homologous non-drosophilid sequences (color-coded as in (D)). The sequence database was assembled using D. melanogaster Gr36a, Grl36a, and Grl43a as the query sequences. Top left: maximum likelihood phylogeny (JTT + F + R7 model) and Bayesian phylogeny. The scale bars represent average number of substitutions per site. Bottom left: phylogenies where weakly supported branches (<95/0.95) have been rearranged and polytomies resolved in a species tree-aware manner. Right: strict consensus of the species tree-aware phylogenies. These analyses support that Gr36 and Grl36a/43a are sister clades, which likely split after Gr59c/d diverged from the ancestral lineage. Sequences are colored as in (D). Select branch support values are present on key branches and refer to maximum likelihood UFboot and Bayesian posterior probabilities, in this order. Asterisks indicate that branch support was below the threshold for species-aware rearrangement. A simplified schematic of gene duplication and loss is illustrated in Figure 3—figure supplement 6F. The fully annotated trees are available in Figure 3—figure supplements 7–9. (F) Histogram of Gr and Grl expression levels in adult proboscis and maxillary palps determined by bulk RNA-sequencing (RNA-seq). Mean values ± SD of fragments per kilobase of transcript per million mapped reads (FPKM) are plotted; n=3 biological replicates. Data is from Dweck et al., 2021. (G) Left: t-distributed stochastic neighbor embedding (tSNE) representation of RNA-seq datasets from individual cells of the D. melanogaster proboscis and maxillary palp – generated as part of the Fly Cell Atlas (10× stringent dataset) (Li et al., 2022) – colored for expression of the indicated genes. Gr64f and Gr66a are broad markers of ‘sweet/appetitive’ and ‘bitter/aversive’ gustatory sensory neurons, respectively. Transcripts for three Grls are detected in subsets of bitter/aversive neurons. Annotations of cell clusters are adapted from Li et al., 2022; unlabeled clusters represent other non-gustatory sensory neuron or non-neuronal cell types of this tissue.
-
Figure 3—source data 1
FASTA file containing the amino acid sequences used in the network and graph splitting analysis of gustatory receptors (Grs), odorant receptors (Ors), and Grls.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data1-v2.zip
-
Figure 3—source data 2
Tab delimited text file containing the sequence similarity network of gustatory receptors (Grs), odorant receptors (Ors), and Grls.
The first column is the source node, the second column is the target node, and the third column is the E-value derived from MMSeqs2 and gs2.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data2-v2.zip
-
Figure 3—source data 3
Tab delimited text file containing the annotation for the sequence similarity network of gustatory receptors (Grs), odorant receptors (Ors), and Grls.
The first column is the node identifier (ID) and the second column is the sequence name (SEQ).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data3-v2.zip
-
Figure 3—source data 4
Newick tree file containing the graph splitting tree of odorant receptors (Ors), gustatory receptors (Grs), and Grls, derived from the sequence similarity network by gs2.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data4-v2.zip
-
Figure 3—source data 5
FASTA file containing the multiple sequence alignment used for illustrating intron and transmembrane domain 7 (TM7) motif conservation between gustatory receptors (Grs) and Grls.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data5-v2.zip
-
Figure 3—source data 6
FASTA file containing the amino acid sequences of Gr36, Gr59, Grl36a, and Grl43a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data6-v2.zip
-
Figure 3—source data 7
FASTA file containing the multiple sequence alignment of Gr36, Gr59, Grl36a, and Grl43a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data7-v2.zip
-
Figure 3—source data 8
Newick tree file containing the maximum likelihood phylogeny of Gr36, Gr59, Grl36a, and Grl43a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data8-v2.zip
-
Figure 3—source data 9
NOTUNG tree file containing the species-aware phylogeny of Gr36, Gr59, Grl36a, and Grl43a homologs, based on the maximum likelihood phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data9-v2.zip
-
Figure 3—source data 10
NEXUS tree file containing the Bayesian phylogeny of Gr36, Gr59, Grl36a, and Grl43a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data10-v2.zip
-
Figure 3—source data 11
NOTUNG tree file containing the species-aware phylogeny of Gr36, Gr59, Grl36a, and Grl43a homologs, based on the Bayesian phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data11-v2.zip
-
Figure 3—source data 12
Newick tree file containing the strict consensus of the species-aware phylogenies of Gr36, Gr59, Grl36a, and Grl43a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-data12-v2.zip
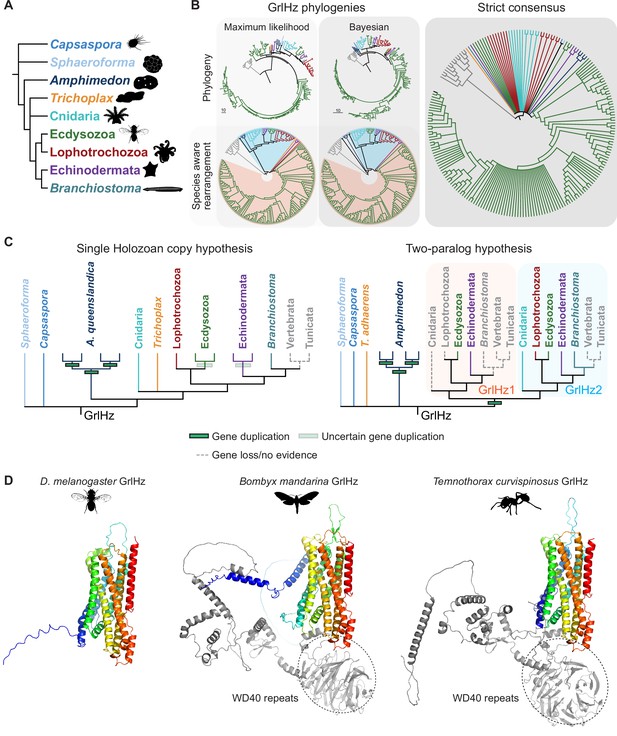
Evolution of GrlHolozoa (GrlHz), a family of Grl seven transmembrane domain ion channel (7TMIC) not restricted to flies.
(A) Major taxa/species for which a GrlHz homolog was recovered. (B) Phylogenies of a representative set of GrlHz sequences (clustered by 70% sequence identity). The sequence database was assembled using D. melanogaster GrlHz as the query sequence. Top: maximum likelihood phylogeny and Bayesian phylogeny. The scale bars represent the average number of substitutions per site. Bottom: phylogenies where weakly supported branches (<95/0.95) have been rearranged and polytomies resolved in a species tree-aware manner. Right: strict consensus of the species tree-aware phylogenies. The fully annotated trees are visualized in Figure 3—figure supplements 2–4. (C) Left: the single holozoan copy hypothesis of GrlHz evolution. Under this scenario, a single GrlHz is widely conserved across Holozoa, but has been independently duplicated/lost several times in various taxa. Right: the two-paralog hypothesis of GrlHz evolution. As both the maximum likelihood and Bayesian phylogenies provide evidence for two GrlHz clades, and because some species have two substantially divergent GrlHz sequences, it is possible that there was a gene duplication event early in the evolution of Holozoa. (D) Examples of GrlHz structures. Of 196 representative sequences, 31 sequences (mostly from Hymenoptera and Lepidoptera) bear N-terminal WD40 repeats.
-
Figure 3—figure supplement 1—source data 1
FASTA file containing the amino acid sequences of validated holozoan GrlHolozoa (GrlHz).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data1-v2.zip
-
Figure 3—figure supplement 1—source data 2
FASTA file containing the representative amino acid sequences of holozoan GrlHolozoa (GrlHz) used in phylogenetic analyses.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data2-v2.zip
-
Figure 3—figure supplement 1—source data 3
FASTA file containing the multiple sequence alignment of holozoan GrlHolozoa (GrlHz).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data3-v2.zip
-
Figure 3—figure supplement 1—source data 4
Newick tree file containing the maximum likelihood phylogeny of holozoan GrlHolozoa (GrlHz).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data4-v2.zip
-
Figure 3—figure supplement 1—source data 5
NOTUNG tree file containing the species-aware phylogeny of holozoan GrlHolozoa (GrlHz), based on the maximum likelihood phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data5-v2.zip
-
Figure 3—figure supplement 1—source data 6
NEXUS tree file containing the Bayesian phylogeny of holozoan GrlHolozoa (GrlHz).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data6-v2.zip
-
Figure 3—figure supplement 1—source data 7
NOTUNG tree file containing the species-aware phylogeny of holozoan GrlHolozoa (GrlHz), based on the Bayesian phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data7-v2.zip
-
Figure 3—figure supplement 1—source data 8
Newick tree file containing the strict consensus of the species-aware phylogenies of holozoan GrlHolozoa (GrlHz).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp1-data8-v2.zip
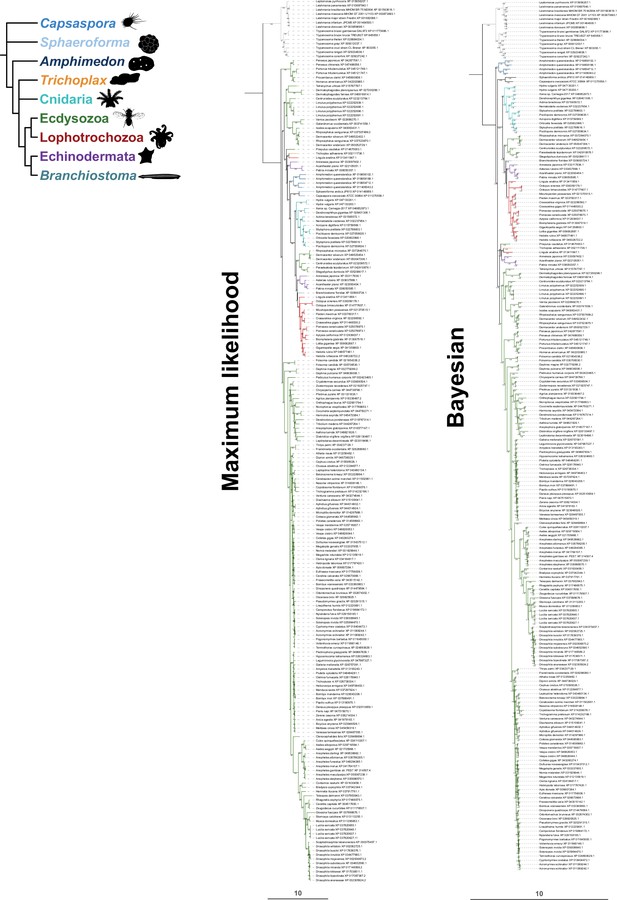
Fully annotated phylogenetic trees for GrlHolozoa (GrlHz) homologs.
For maximum likelihood, the tree was generated using a JTT + F + R9 substitution model. Branch support values for maximum likelihood (UFboot) and Bayesian analyses (posterior probability) are shown at the branches. The scale bars represent the average number of substitutions per site.
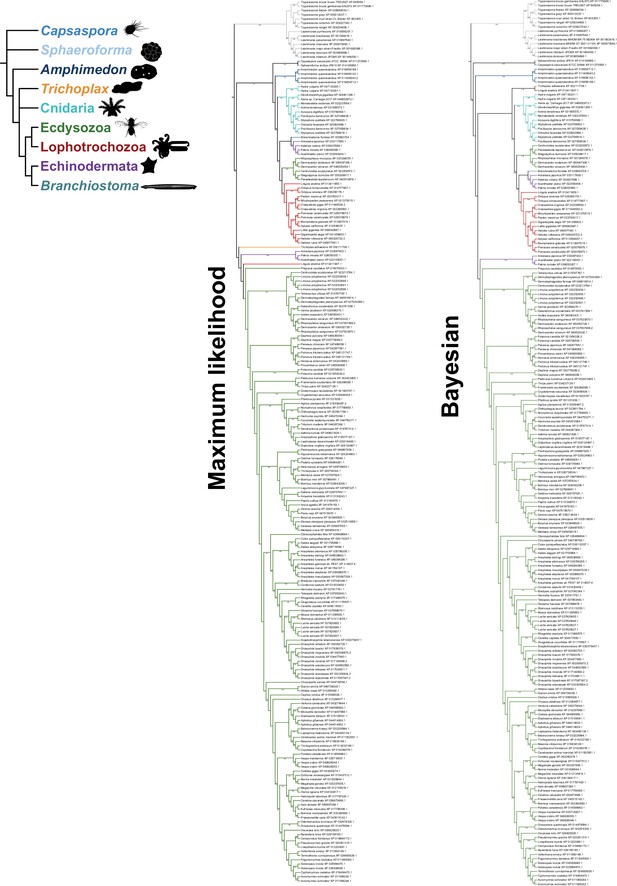
Fully annotated species-aware trees for GrlHolozoa (GrlHz) homologs.
Trees are based on the maximum likelihood (left) and Bayesian (right) trees. Branches without support values were eligible for rearrangement.
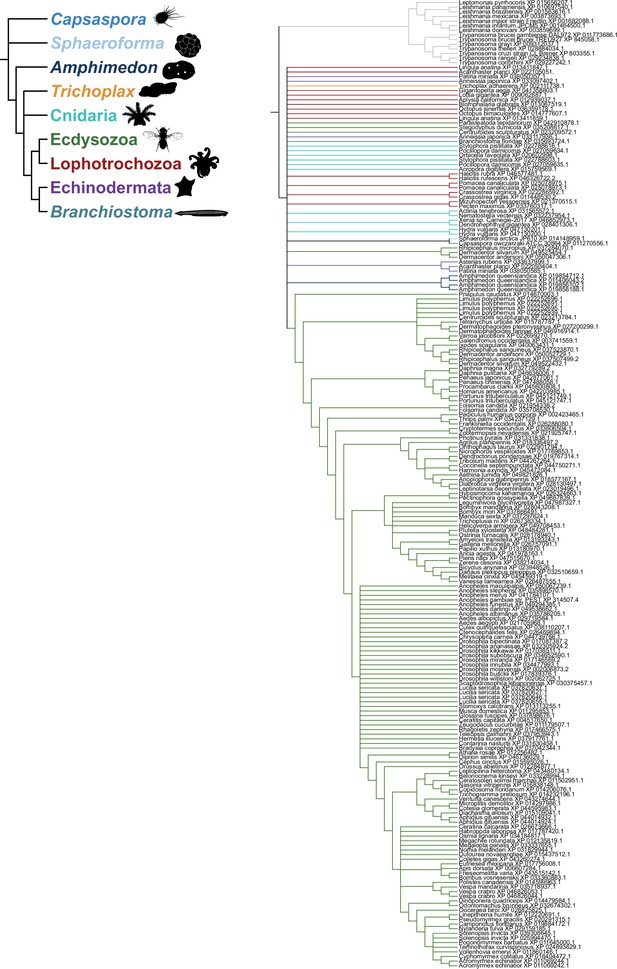
Strict consensus of the species-aware trees for GrlHolozoa (GrlHz) homologs.
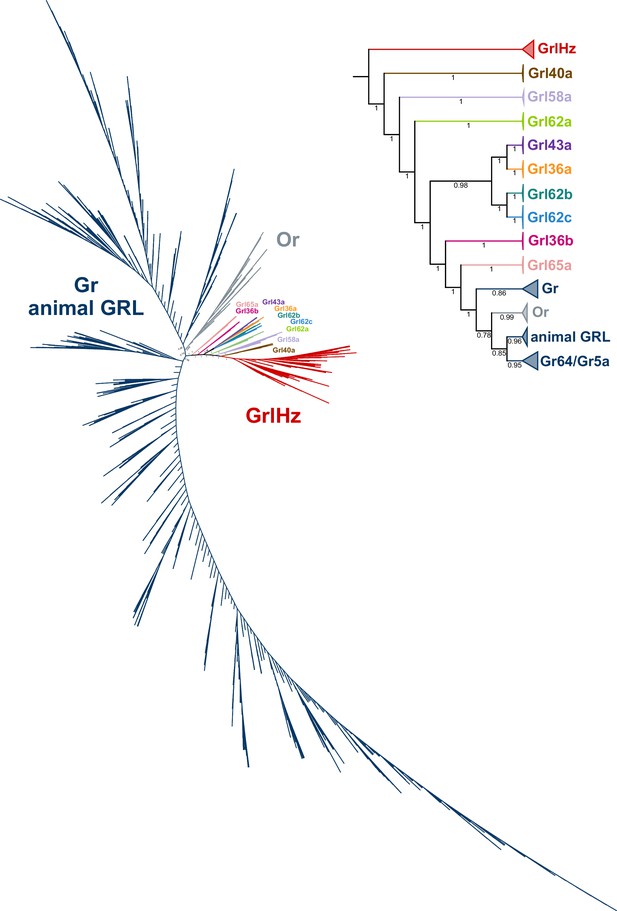
Fully annotated graph splitting tree for odorant receptors (Ors), gustatory receptors (Grs), and Grls.
Key edge perturbation support values are visible on branches. The primary sequence databases were assembled using each of the D. melanogaster Grls as query sequences. D. melanogaster Or and Gr sequences were manually collected from FlyBase. Sequences from M. hrabei (jumping bristletail), Thermobia domestica (firebrat), Ladona filva (dragonfly), and Ephemera danica (green drake mayfly) were added, following the proposal that canonical Ors may have diversified after the emergence of Neoptera (most winged insects) (Brand et al., 2018); 2498 additional sequences were collected using the N. vectensis GRL1 query sequence (XP_048580785.1); the PSI-BLAST searches were stopped at four iterations, as the search had substantially recovered insect Gr sequences, and further searches returned tens of thousands of sequences. The basal placement of the Grls is unusual given their conservation in flies, as this would suggest they diversified in a common animal ancestor and that the Grls were lost in all animal taxa except flies. This hypothesis seems unlikely given the extreme number of independent gene loss events this would require, and we therefore suspect that this tree topology represents a phylogenetic error, for example, long branch attraction (Bergsten, 2005). The inset shows major collapsed clades, where the tip node is sized proportionally to the number of sequences collapsed.
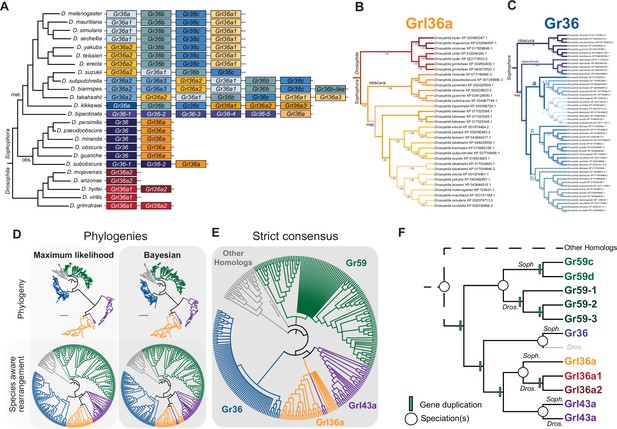
The evolution of Gr36, Gr59, Grl36a, and Grl43a.
(A) Schematic of the gene arrangement of Grl36a and Gr36 homologs in drosophilids, with colors matching trees in (B) and (C). This panel is reproduced from Figure 3C. (B) Species-aware Bayesian phylogeny of Grl36. (C) Species tree-aware Bayesian phylogeny of Gr36. (D) Phylogenies of Gr36, Gr59, Grl36a, Grl43a, and other homologous sequences. The sequence database was assembled using D. melanogaster Gr36a, Grl36a, and Grl43a as query sequences. Top: maximum likelihood phylogeny and Bayesian phylogeny. The scale bars represent the average number of substitutions per site. Bottom: phylogenies where weakly supported branches (<95/0.95) have been rearranged and polytomies resolved in a species tree-aware manner. (E) Strict consensus of the species tree-aware phylogenies. These analyses support that Gr36 and Grl36a/43a are sister clades, which likely split after the Gr59 split. (F) Proposed model of Gr36, Gr59, Grl36a, and Grl43a evolution.
-
Figure 3—figure supplement 6—source data 1
FASTA file containing the amino acid sequences of Gr36 homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data1-v2.zip
-
Figure 3—figure supplement 6—source data 2
FASTA file containing the multiple sequence alignment of Gr36 homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data2-v2.zip
-
Figure 3—figure supplement 6—source data 3
Newick tree file containing the maximum likelihood phylogeny of Gr36 homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data3-v2.zip
-
Figure 3—figure supplement 6—source data 4
NOTUNG tree file containing the species-aware phylogeny of Gr36 homologs, based on the maximum likelihood phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data4-v2.zip
-
Figure 3—figure supplement 6—source data 5
NEXUS tree file containing the Bayesian phylogeny of Gr36 homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data5-v2.zip
-
Figure 3—figure supplement 6—source data 6
NOTUNG tree file containing the species-aware phylogeny of Gr36 homologs, based on the Bayesian phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data6-v2.zip
-
Figure 3—figure supplement 6—source data 7
FASTA file containing the amino acid sequences of Grl36a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data7-v2.zip
-
Figure 3—figure supplement 6—source data 8
FASTA file containing the multiple sequence alignment of Grl36a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data8-v2.zip
-
Figure 3—figure supplement 6—source data 9
Newick tree file containing the maximum likelihood phylogeny of Grl36a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data9-v2.zip
-
Figure 3—figure supplement 6—source data 10
NOTUNG tree file containing the species-aware phylogeny of Grl36a homologs, based on the maximum likelihood phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data10-v2.zip
-
Figure 3—figure supplement 6—source data 11
NEXUS tree file containing the Bayesian phylogeny of Grl36a homologs.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data11-v2.zip
-
Figure 3—figure supplement 6—source data 12
NOTUNG tree file containing the species-aware phylogeny of Grl36a homologs, based on the Bayesian phylogeny.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig3-figsupp6-data12-v2.zip
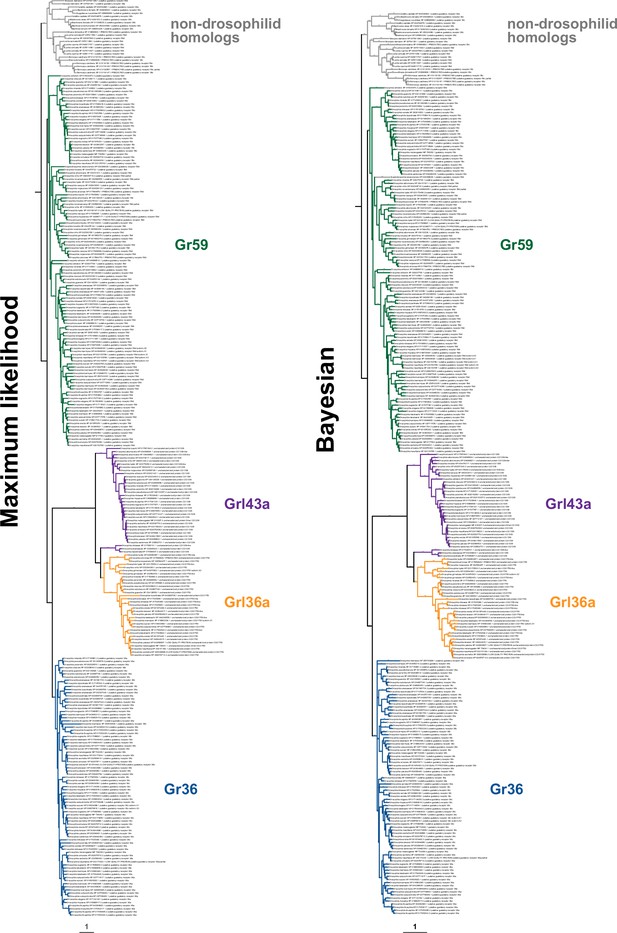
Fully annotated phylogenetic trees for Gr36, Gr59, Grl36a, and Grl43a homologs.
For maximum likelihood, the tree was generated using a JTT + F + R7 substitution model and is rooted. Branch support values for maximum likelihood (UFboot) and Bayesian analyses (posterior probability) are shown at the branches. Non-drosophilid sequences are assumed to be the outgroup. The scale bars represent the average number of substitutions per site.
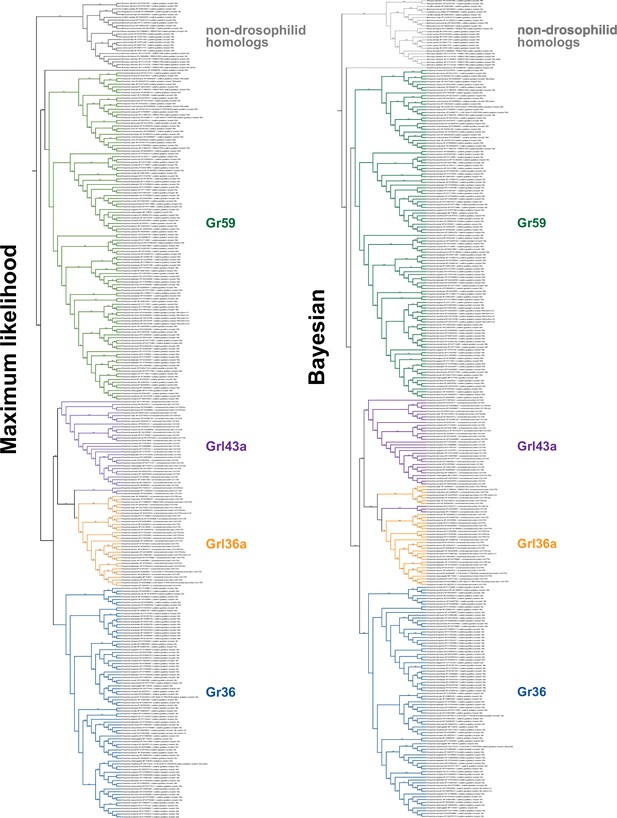
Fully annotated species-aware trees for Gr36, Gr59, Grl36a, and Grl43a homologs.
Trees are based on the maximum likelihood (left) and Bayesian (right) trees. Branches without support values were eligible for rearrangement.
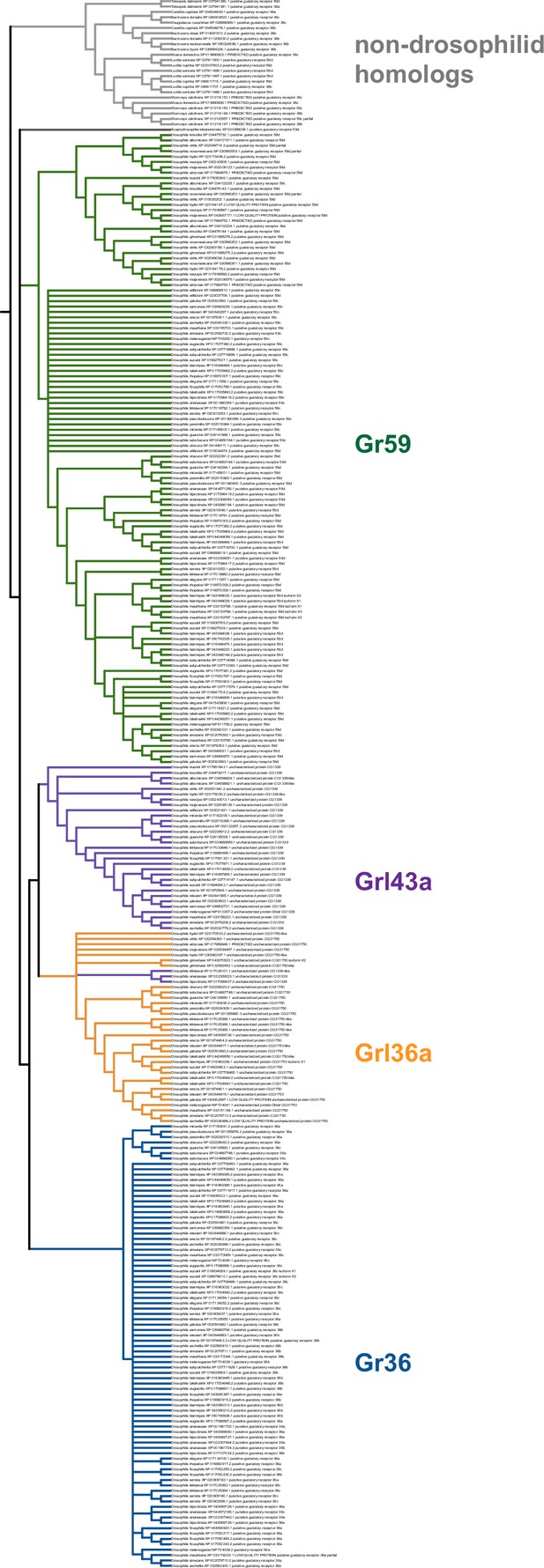
Strict consensus of the species-aware trees for Gr36, Gr59, Grl36a, and Grl43a homologs.
Although the consensus tree has a polytomy near the emergence of Gr59, this is strictly due to disagreement as to whether the lone Scaptodrosophila sequence is a Gr59 homolog or an outgroup to all other Drosophila/Sophophora sequences shown here.
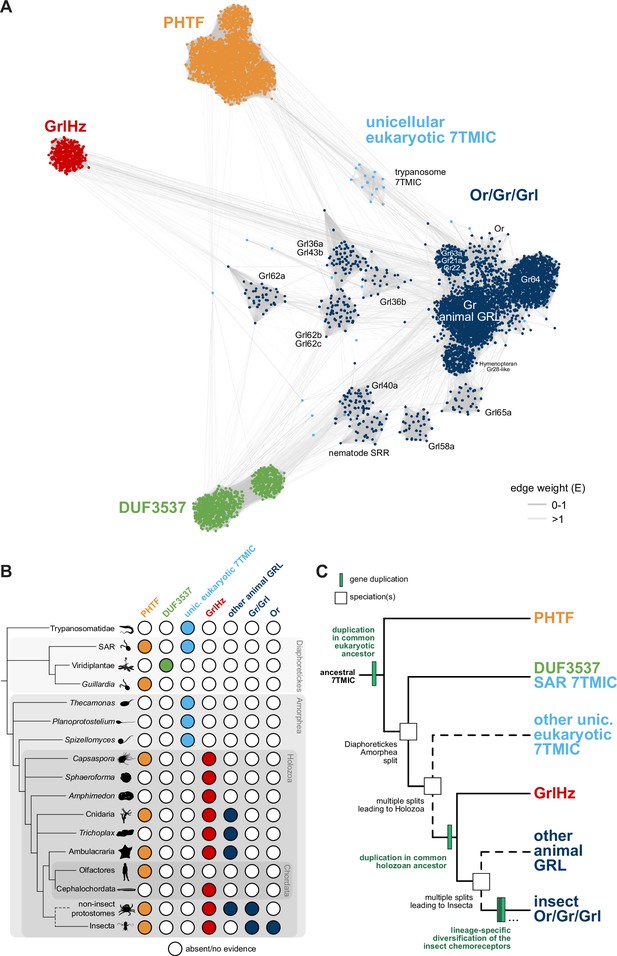
A hypothesis for the evolution of the seven transmembrane domain ion channel (7TMIC) superfamily.
(A) Sequence similarity network of the 7TMIC superfamily, generated using the same odorant receptors (Ors) and gustatory receptors (Grs) from Figure 3B, unicellular eukaryotic Grls from Benton et al., 2020, and sequence databases assembled using the following query sequences: N. vectensis GRL1, D. melanogaster Grls and Phtf, H. sapiens PHTF1 and PHTF2, Arabidopsis thaliana Domain of Unknown Function (DUF) 3537, C. elegans SRRs and trypanosome GRLs. The network was generated and visualized as in Figure 3B. The graph splitting tree is visualized in Figure 4—figure supplement 1. (B) Presence and absence of 7TMICs across taxa: ‘other animal GRL’ refers to GRLs in non-insect animal species previously identified by primary sequence similarity (Benton, 2015; Robertson, 2015; Saina et al., 2015) and nematode SRRs. The dashed branch represents several collapsed paraphyletic clades. (C) Model of 7TMIC superfamily evolution. The dashed branches represent several collapsed paraphyletic clades and speciation events. The trypanosome 7TMICs are unplaced due to the currently unresolved taxonomy of trypanosomes (Burki et al., 2020).
-
Figure 4—source data 1
FASTA file containing the amino acid sequences used in the network and graph splitting analysis of eukaryotic seven transmembrane domain ion channels (7TMICs).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig4-data1-v2.zip
-
Figure 4—source data 2
Tab delimited text file containing the sequence similarity network of eukaryotic seven transmembrane domain ion channels (7TMICs).
The first column is the source node, the second column is the target node, and the third column is the E-value derived from MMSeqs2 and gs2.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig4-data2-v2.zip
-
Figure 4—source data 3
Tab delimited text file containing the annotation for the sequence similarity network of eukaryotic seven transmembrane domain ion channels (7TMICs).
The first column is the node identifier (ID) and the second column is the sequence name (SEQ).
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig4-data3-v2.zip
-
Figure 4—source data 4
Newick tree file containing the graph splitting tree of eukaryotic seven transmembrane domain ion channels (7TMICs), derived from the sequence similarity network by gs2.
- https://cdn.elifesciences.org/articles/85537/elife-85537-fig4-data4-v2.zip
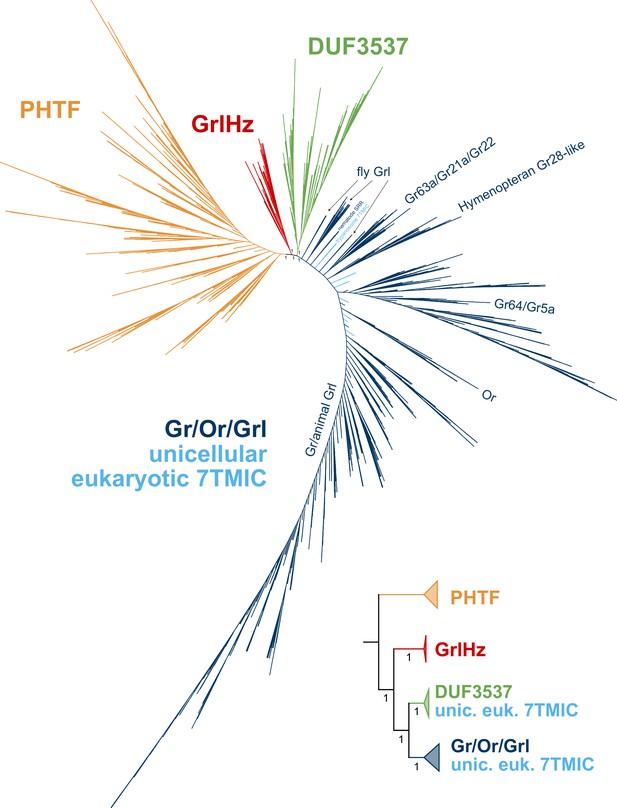
Graph splitting tree for the proposed seven transmembrane domain ion channel (7TMIC) superfamily.
Key edge perturbation support values are visible on branches. The inset shows major collapsed clades, where the triangular tip is sized proportionally to the number of sequences collapsed. This tree suggests a different branching pattern than the hypothesis in Figure 4C, consistent with a more complex duplication/loss history for the 7TMIC superfamily. However, as in Figure 3—figure supplement 5, we suspect long branch attraction is present in this analysis, at least for the fly Grls and nematode proteins.
Tables
Quantitative structural comparisons of candidate seven transmembrane domain ion channel (7TMIC) homologs.
Summary of amino acid identity (%), Dali Z-score, and TM-align TM-score of the indicated experimentally determined or ab initio-predicted structures of 7TMIC homologs (or negative-control, unrelated proteins) compared to A. bakeri Or co-receptor (Orco). The Orco cryo-electronic microscopic (cryo-EM) structure chain A (6C70-A) (Butterwick et al., 2018) was used as the query in all comparisons. Protein models are provided in Source data 1. Note the nomenclature of unicellular eukaryotic 7TMICs is tentative; identical names (e.g., GRL1) do not imply orthology. Typically, a Z-score >20 indicates that the two proteins being compared are definitely homologous, 8–20 that they are probably homologous, and 2–8 is a ‘gray area’ influenced by protein size and fold (Holm, 2020). TM-scores of 0.5–1 indicate that the two proteins being compared adopt generally the same fold, while TM-scores of 0–0.3 indicate random structural similarity (Zhang and Skolnick, 2004; Zhang and Skolnick, 2005). For the negative controls, the amino acid identity differs slightly between the experimentally determined and ab initio-predicted proteins because of small differences in sequence coverage.
| Category | Protein | Model or PDB | Method or algorithm | Comparison to A. bakeri Orco (6C70-A) | ||
|---|---|---|---|---|---|---|
| Amino acid identity (%) | DaliZ-score | TM-alignTM-score | ||||
| Positive controls (known 7TMIC) | A. bakeri Orco | 61b81_unrelaxed_rank_1_model_2 | AlphaFold2 | 100 | 50.7 | 0.96 |
| M. hrabei Or5 | 7LIC-A | Cryo-EM | 19 | 36.3 | 0.81 | |
| Drosophila melanogaster Gr64a | AF-P83293-F1-model_v4 | AlphaFold2 | 13 | 29.6 | 0.79 | |
| N. vectensis GRL1 | AF-A7S7G0-F1-model_v4 | AlphaFold2 | 10 | 31.3 | 0.78 | |
| Unicellular eukaryotic 7TMIC | Thecamonas trahens GRL1 | AF-A0A0L0DUY0-F1-model_v3 | AlphaFold2 | 9 | 23.2 | 0.71 |
| T. trahens GRL2 | AF-A0A0L0DQC1-F1-model_v3 | AlphaFold2 | 12 | 25.3 | 0.70 | |
| T. trahens GRL3 | AF-A0A0L0D5B5-F1-model_v3 | AlphaFold2 | 14 | 13.1 | 0.50 | |
| T. trahens GRL4 | AF-A0A0L0D5H0-F1-model_v3 | AlphaFold2 | 9 | 9.9 | 0.53 | |
| T. trahens GRL5 | AF-A0A0L0DD38-F1-model_v3 | AlphaFold2 | 10 | 12.2 | 0.56 | |
| T. trahens GRL6 | AF-A0A0L0DJ52-F1-model_v3 | AlphaFold2 | 8 | 15.6 | 0.57 | |
| V. brassicaformis GRL1 | AF-A0A0G4FIT4-F1-model_v3 | AlphaFold2 | 10 | 9.1 | 0.47 | |
| V. brassicaformis GRL2 | AF-A0A0G4ECU2-F1-model_v3 | AlphaFold2 | 11 | 14.4 | 0.57 | |
| V. brassicaformis GRL3 | AF-A0A0G4FWI7-F1-model_v3 | AlphaFold2 | 14 | 23.8 | 0.74 | |
| V. brassicaformis GRL4 | AF-A0A0G4EU86-F1-model_v3 | AlphaFold2 | 10 | 18.5 | 0.70 | |
| V. brassicaformis GRL5 | AF-A0A0G4FBY6-F1-model_v3 | AlphaFold2 | 10 | 18.5 | 0.68 | |
| V. brassicaformis GRL6 | AF-A0A0G4G8W6-F1-model_v3 | AlphaFold2 | 8 | 21.4 | 0.70 | |
| Micromonas pusilla GRL1 | AF-C1MGH9-F1-model_v3 | AlphaFold2 | 12 | 11.3 | 0.60 | |
| Chloropicon primus GRL1 | AF-A0A5B8MFA4-F1-model_v3 | AlphaFold2 | 10 | 18.1 | 0.71 | |
| L. infantum GRL1 | AF-A4HWQ9-F1-model_v3 | AlphaFold2 | 6 | 13.5 | 0.64 | |
| T. brucei GRL1 | AF-Q57U78-F1-model_v3 | AlphaFold2 | 9 | 13.4 | 0.62 | |
| Fly Grl | D. melanogaster Grl36a | AF-Q8INZ1-F1-model_v3 | AlphaFold2 | 9 | 19.5 | 0.67 |
| D. melanogaster Grl36b | AF-Q8INY2-F1-model_v3 | AlphaFold2 | 8 | 15.2 | 0.62 | |
| D. melanogaster Grl40a | AF-Q0E8M7-F1-model_v3 | AlphaFold2 | 8 | 19.5 | 0.66 | |
| D. melanogaster Grl43a | AF-Q9V4Q0-F1-model_v3 | AlphaFold2 | 10 | 19.9 | 0.69 | |
| D. melanogaster Grl58a | AF-Q9W2A4-F1-model_v3 | AlphaFold2 | 8 | 15.0 | 0.60 | |
| D. melanogaster Grl62a | AF-B7Z0I0-F1-model_v3 | AlphaFold2 | 8 | 19.4 | 0.69 | |
| D. melanogaster Grl62b | AF-B7Z0I1-F1-model_v3 | AlphaFold2 | 11 | 19.1 | 0.66 | |
| D. melanogaster Grl62c | AF-Q6ILZ2-F1-model_v3 | AlphaFold2 | 10 | 17.2 | 0.63 | |
| D. melanogaster Grl65a | AF-Q8IQ72-F1-model_v3 | AlphaFold2 | 11 | 25.9 | 0.74 | |
| D. melanogaster GrlHz | AF-Q9W1W8-F1-model_v3 | AlphaFold2 | 7 | 22.5 | 0.74 | |
| PHTF | Homo sapiens PHTF1 | AF-Q9UMS5-F1-model_v3 | AlphaFold2 | 7 | 12.9 | 0.63 |
| H. sapiens PHTF2 | AF-Q8N3S3-F1-model_v3 | AlphaFold2 | 8 | 12.0 | 0.62 | |
| D. melanogaster Phtf | AF-Q9V9A8-F1-model_v3 | AlphaFold2 | 5 | 11.8 | 0.63 | |
| Negative controls (non-7TMIC) | Bos taurus Rhodopsin | 1F88-A | X-ray crystal | 9 | 2.1 | 0.31 |
| AF-P02699-F1-model_v4 | AlphaFold2 | 9 | <2.0 | 0.19 | ||
| Chlamydomonas reinhardtii ChR2 | 6EID-A | X-ray crystal | 7 | 3.6 | 0.27 | |
| AF-Q8RUT8-F1-model_v4 | AlphaFold2 | 9 | 3.4 | 0.10 | ||
| H. sapiens Frizzled4 | 6BD4 | X-ray crystal | 8 | 4.0 | 0.34 | |
| AF-Q9ULV1-F1-model_v4 | AlphaFold2 | 5 | 2.9 | 0.19 | ||
| H. sapiens AdipR | 5LXG | X-ray crystal | 2 | 3.6 | 0.29 | |
| AF-Q96A54-F1-model_v4 | AlphaFold2 | 2 | <2.0 | 0.14 | ||
| Escherichia coli GlpG | 2XOV | X-ray crystal | 5 | 3.5 | 0.27 | |
| AF-P09391-F1-model_v4 | AlphaFold2 | 6 | 3.3 | 0.13 | ||
| Mus musculus TRPV3 | 6LGP-D | Cryo-EM | 10 | 2.7 | 0.27 | |
| AF-Q8K424-F1-model_v4 | AlphaFold2 | 14 | 2.3 | 0.08 | ||
| M. musculus Piezo | 6BPZ-B | Cryo-EM | 5 | 4.0 | 0.27 | |
| AF-E2JF22-F1-model_v4 | AlphaFold2 | 5 | 2.3 | 0.08 | ||
| B. taurus CNGA/CNGB | 7O4H-A | Cryo-EM | 9 | 2.8 | 0.24 | |
| AF-Q00194-F1-model_v4 | AlphaFold2 | 9 | 3.3 | 0.11 | ||
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85537/elife-85537-mdarchecklist1-v2.pdf
-
Source data 1
AlphaFold2 models.
Models of proteins analyzed in this work, either downloaded from the AlphaFold Protein Structure Database or, where not already available, predicted using the AlphaFold2 algorithm implemented in ColabFold (Mirdita et al., 2022). The four-letter code in the filename represents the first letter of the genus and the first three letters of the species (e.g., ‘Dmel’ = D. melanogaster); species names are given in full in the figures.
- https://cdn.elifesciences.org/articles/85537/elife-85537-data1-v2.zip
-
Source data 2
Dali screen search results.
Individual text files represent the output of the Dali AF-DB search using A. bakeri Or co-receptor (Orco) chain A (PDB 6C70-A) as query and the structural proteome dataset of the indicated species (note the datasets are from version 1 of the AlphaFold Protein Structure Database; subsequent, improved models were used for the pairwise comparisons in Table 1). The four-letter codes in the file names and job titles are as described for Source data 1.
- https://cdn.elifesciences.org/articles/85537/elife-85537-data2-v2.zip
-
Source data 3
Reverse Dali search results.
Individual text files represent the output of the Dali AF-DB search using the indicated query candidate seven transmembrane domain ion channels (7TMICs) from Trypanosoma (GRL1), D. melanogaster (Grls), or H. sapiens (PHTF1/2) and the structural proteomic dataset of D. melanogaster.
- https://cdn.elifesciences.org/articles/85537/elife-85537-data3-v2.zip
-
Source data 4
All uncurated PSI-BLAST sequence databases.
Each of the FASTA filenames is formatted as follows (with the exception of the D. melanogaster odorant receptor (Or) and gustatory receptor (Gr) sequences, which were collected manually from FlyBase): ProteinFamily-QuerySpecies-QuerySequence.fasta.
- https://cdn.elifesciences.org/articles/85537/elife-85537-data4-v2.zip