Inhibition of DNMT1 methyltransferase activity via glucose-regulated O-GlcNAcylation alters the epigenome
Figures
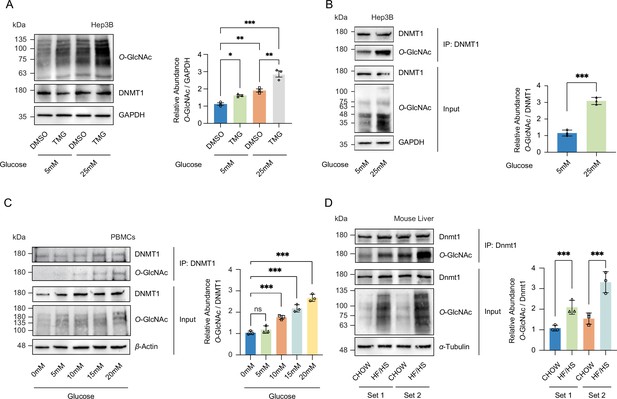
High glucose increases O-GlcNAcylation of DNMT1 in cell lines and primary cells.
(A) Hep3B cells were treated with glucose (5 mM or 25 mM) with or without Thiamet-G (TMG). Shown are immunoblots of collected lysates using antibody targeting O-GlcNAc and GAPDH (n = 3). (B) Lysates of Hep3B treated with glucose were immunoprecipitated with DNMT1 and immunoprecipitates were immunoblotted with antibody targeting O-GlcNAc (n = 3). (C) Peripheral blood mononuclear cells (PBMCs) were isolated from three individual donor blood samples and treated with increasing concentration of glucose for 24 hr. Collected cell lysates from PBMCs were immunoprecipitated with antibody targeting DNMT1 and immunoblotted for O-GlcNAc. Representative blot from one donor (n = 3). (D) Immunoblots for O-GlcNAc and GAPDH from liver samples of C57BL/6J mice given a high-fat/high-sucrose diet (HF/HS) or normal diet (chow) for 4 mo, and immunoprecipitated with Dnmt1. Lysates of mouse liver were immunoprecipitated with Dnmt1 and immunoprecipitates were immunoblotted with antibody targeting O-GlcNAc. *p<0.001; **p<0.0005; ***p<0.0001 by Student’s t-test (A-D); ns, not significant; data are represented as mean ± SD from three replicates of each sample.
-
Figure 1—source data 1
Uncropped blot files of Figure 1A–D.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig1-data1-v2.zip
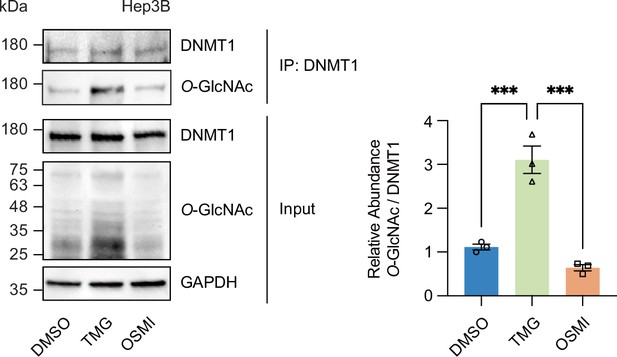
DNMT1 can be O-GlcNAcylated in Hep3B cells.
Hep3B cells were treated with Thiamet-G (TMG) or OSMI-4 (OSMI). Shown are representative immunoblots of treated Hep3B lysates performed with antibodies targeting O-GlcNAc and GAPDH and bar graphs of relative expression between O-GlcNAc compared to control, GAPDH (n = 3, experimental replicates). Lysates from treated Hep3B with glucose were immunoprecipitated with DNMT1 and immunoprecipitates were immunoblotted with antibody targeting O-GlcNAc (n = 3). ***p<0.0001 by Student’s t-test; Data are represented as mean ± SD from three replicates of each sample.
-
Figure 1—figure supplement 1—source data 1
Uncropped blot files of Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig1-figsupp1-data1-v2.zip
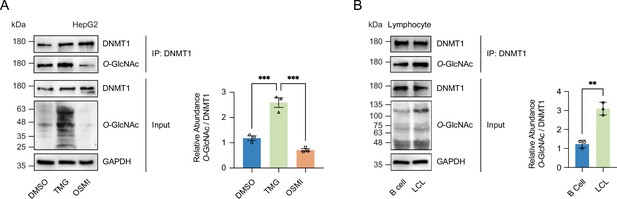
DNMT1 can be O-GlcNAcylated in HepG2 cells and B-cells-derived lymphocytes.
(A) HepG2 cells were treated with Thiamet-G or OSMI. Shown are immunoblots of treated HepG2 lysates performed with immunoblots of immunoprecipitates performed with antibodies targeting O-GlcNAc (n = 3). (B) Shown are immunoblots of B cell and lymphocytes (LCL) lysates performed with immunoblots of immunoprecipitates performed with antibodies targeting O-GlcNAc (n = 3). **p<0.0005, ***p<0.0001 by Student’s t-test (A, B); data are represented as mean ± SD from three replicates of each sample.
-
Figure 1—figure supplement 2—source data 1
Uncropped blot files of Figure 1—figure supplement 2A and B.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig1-figsupp2-data1-v2.zip
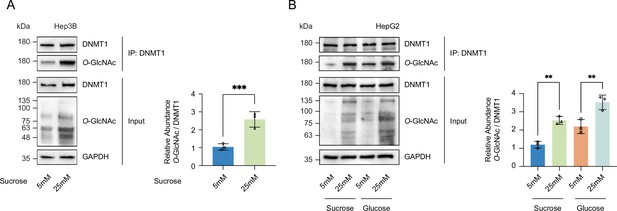
Global protein O-GlcNAcylation was induced with high concentrations of sucrose.
(A) Hep3B cells were treated with sucrose (5 mM or 25 mM). Shown are immunoblots of collected lysates using antibody targeting O-GlcNAc and GAPDH (n = 3). Lysates of Hep3B treated with sucrose were immunoprecipitated with DNMT1 and immunoprecipitates were immunoblotted with antibody targeting O-GlcNAc (n = 3). (B) HepG2 cells were treated with 5 mM glucose or sucrose, or 25 mM glucose or sucrose. Lysates of HepG2 treated with glucose were immunoprecipitated with DNMT1 and immunoprecipitates were immunoblotted with antibody targeting O-GlcNAc (n = 3). **p<0.0005; ***p<0.0001 by Student’s t-test (A, B); data are represented as mean ± SD from three replicates of each sample.
-
Figure 1—figure supplement 3—source data 1
Uncropped blot files of Figure 1—figure supplement 3A and B.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig1-figsupp3-data1-v2.zip
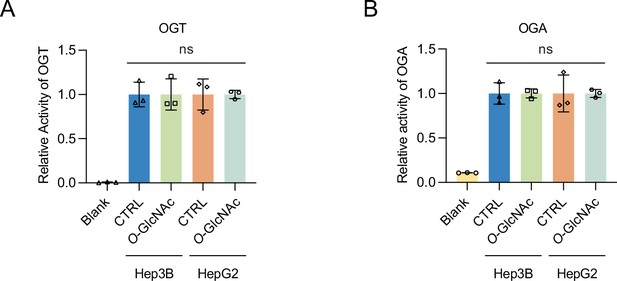
The enzymatic activity of OGT or OGA was not significantly changed by glucose treatment.
(A) OGT activity was measured with low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) using the UDP-Glo Glycosyltransferase activity kit (Promega) (n = 3). (B) OGA activity was measured with low (5 mM, CTRL) or high glucose with TMG (25 mM, O-GlcNAc) using the O-GlcNAcase (OGA, NAG, or MGEA5) assay kit (Biomedical Research Service & Clinical Application) (n = 3). ns, not significant; data are represented as mean ± SD from three replicates of each sample.
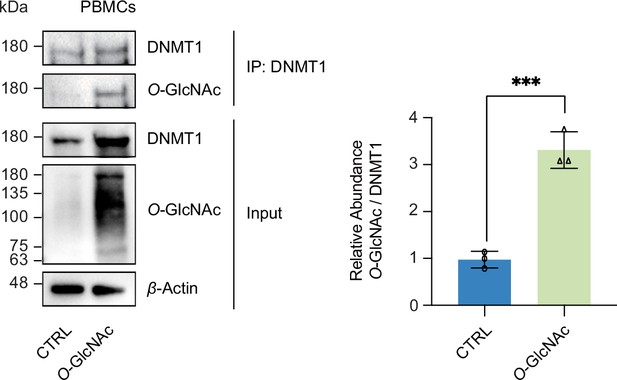
DNMT1 can be O-GlcNAcylated in primary cells (peripheral blood mononuclear cells [PBMCs]).
Pooled PBMCs treated with low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) were immunoprecipitated with antibody targeting DNMT1 and immunoblotted for O-GlcNAc (n = 3). ***p<0.0001 by Student’s t-test; data are represented as mean ± SD from three replicates of each sample.
-
Figure 1—figure supplement 5—source data 1
Uncropped blot files of Figure 1—figure supplement 5.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig1-figsupp5-data1-v2.zip
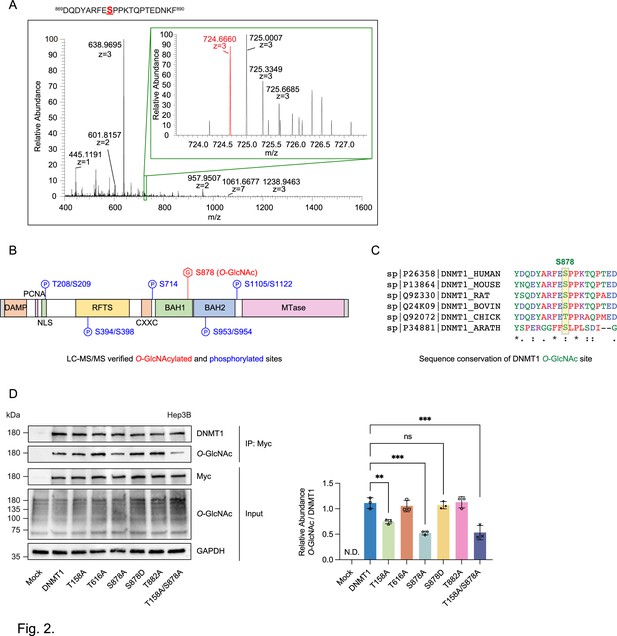
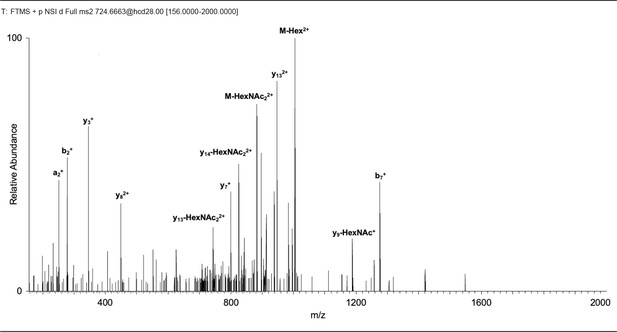
Identification of O-GlcNAcylated sites within DNMT1 by LC-MS/MS.
(A) Schematic drawing of the DNMT1 O-GlcNAc-modified region enriched from Hep3B cells based on mass spectrometry (MS) data and tandem MS (MS/MS) peaks. FTMS+ p NSI full MS (400.0000–1600.0000). DQDYARFESPPKTQPTEDNKF (S9 HexNAc) – S878. (B) Schematic diagram of identified novel O-GlcNAcylated and phosphorylated sites within DNMT1 as determined via LC-MS/MS. DMAP, DNA methyltransferase associated protein-binding domain; PCNA, proliferating cell nuclear antigen-binding domain; NLS, nuclear localization sequences; RFTS, replication foci targeting sequence domain; BAH, bromo-adjacent homology domain. (C) Sequence conservation of S878 in vertebrates. (D) Each immunoprecipitated Myc-DNMT1 wild type and substituted mutants was immunoblotted with an O-GlcNAc antibody (n = 3). **p<0.0005; ***p<0.0001 by Student’s t-test (D); N.D., not detected, ns, not significant; data are represented as mean ± SD from three replicates of each sample.
-
Figure 2—source data 1
Uncropped blot files of Figure 2D.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig2-data1-v2.zip
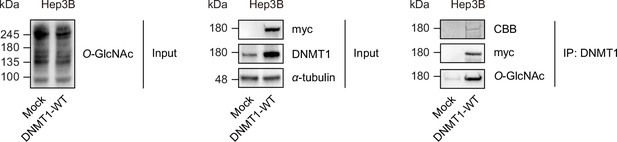
Myc-DNMT1-WT in Hep3B cells can be O-GlcNAcylated.
Myc-DNMT1-WT were transfected into Hep3B cells. Shown are immunoblots of treated DNMT1-WT lysates performed with antibodies targeting Myc, DNMT1, Tubulin, H3, and O-GlcNAc (n = 3). Lysates from treated DNMT1-WT were immunoprecipitated with Myc antibody. Shown are immunoblots of immunoprecipitates performed with antibodies targeting O-GlcNAc and CBB stained gel (Coomassie Brilliant Blue stain).
-
Figure 2—figure supplement 1—source data 1
Uncropped blot files of Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig2-figsupp1-data1-v2.zip
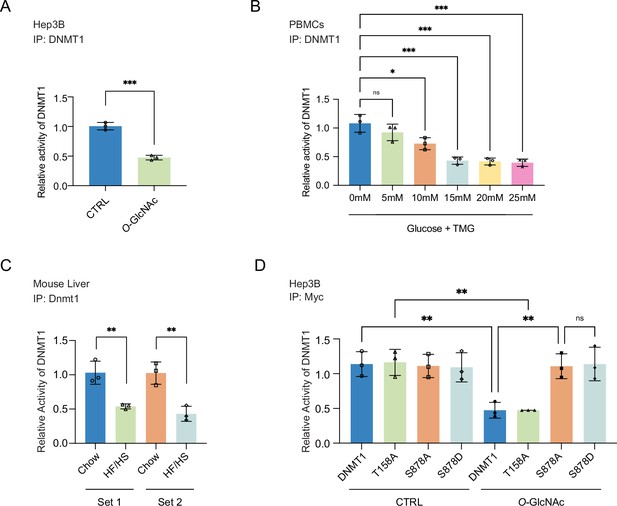
Site-specific O-GlcNAcylation inhibits DNMT1 methyltransferase function.
For (A–D), bar graphs are of relative activity of DNA methyltransferase activity measured as absorbance from a DNMT activity/Inhibition ELISA kit and representative immunoblots of immunoprecipitates performed with antibodies targeting DNMT1. (A) Hep3B cells were treated with low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) (n = 3). (B) Peripheral blood mononuclear cells (PBMCs) from donors were treated with increasing concentrations of glucose (range: 0–25 mM with TMG) (n = 3). (C) Liver samples from C57BL/6J mice given a high-fat/high-sucrose diet (HF/HS) or a normal diet (chow) for 4 mo. (D) Immunoprecipitated DNMT1 wild type and substituted mutants were treated with 5 mM or 25 mM glucose (n = 3). *p<0.001; **p<0.0005; ***p<0.0001 by Student’s t-test (A–D); ns, not significant; data are represented as mean ± SD from three replicates of each sample.
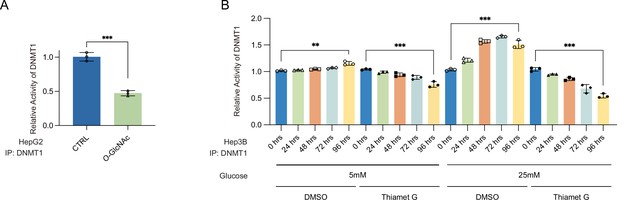
Site-specific O-GlcNAcylation at DNMT1 sites abrogate the function of methyltransferase and DNA loss of methylation at CpG island under high glucose/Thiamet-G (TMG) conditions.
(A) HepG2 cells were treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). Shown are absorbance of DNA methyltransferase activity performed with DNA methyltransferase activity kit (n = 3, technical replicates from three biological replicates for each strain). (B) Hep3B cells were treated with 25 mM glucose with or without Thiamet-G by time dependent. Shown are absorbance of DNA methyltransferase activity performed with DNA methyltransferase activity kit (n = 3, technical replicates from three biological replicates for each strain). **p<0.0005, ***p<0.0001 by Student’s t-test (A, B); data are represented as mean ± SD from three replicates of each sample.
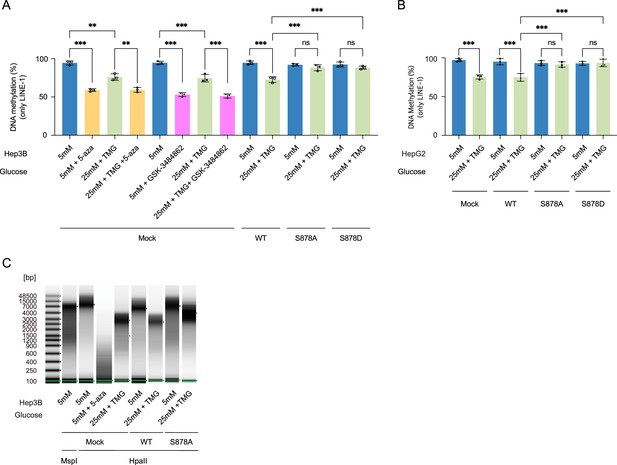
The methylation loss by high glucose/Thiamet-G (TMG) conditions was not apparent in the DNMT1-S878A mutant.
(A) Each Hep3B and Myc-DNMT1-overexpressed mutant (DNMT1-WT or DNMT1-S878A) was treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc), 5-aza or GSK-3484862 (negative control). Shown are absorbance of global DNA methylation of LINE-1 performed with global DNA methylation LINE-1 kit (n = 3). (B) Each HepG2 and Myc-DNMT1-overexpressed mutants (DNMT1-WT or DNMT1-S878A) were treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). Shown are absorbance of global DNA methylation of LINE-1 performed with global DNA methylation LINE-1 kit (n = 3, technical replicates from three biological replicates for each strain). (C) DNA was extracted from Hep3B and Myc-DNMT1-overexpressed mutants (DNMT1-WT or DNMT1-S878A) were treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc) or 5-aza (negative control) with MspI (negative control) or HpaII. Shown are extracted genomic DNA samples and analyze on the 4200 TapeStation System with the Genomic DNA Screen Tape assay with methylation-sensitive enzyme using MspI or HpaII (n = 3, technical replicates from three biological replicates for each strain). **p<0.0005; ***p<0.0001 by Student’s t-test (A, B); ns, not significant; data are represented as mean ± SD from three replicates of each sample.
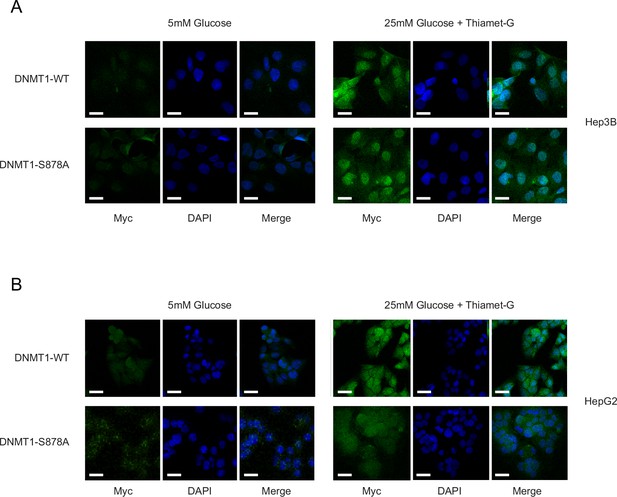
High glucose/Thiamet-G (TMG) conditions do not impact localization of DNMT1-WT or DNMT1-S878A.
(A) Hep3B cells expressed with DNMT1-WT and DNMT1-S878 cells were treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). (B) HepG2 cells expressed with DNMT1-WT and DNMT1-S878 cells were treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). DNMT1 localization was determined by immunofluorescence using confocal microscopy.
-
Figure 3—figure supplement 3—source data 1
Raw fluorescence image files of Figure 3.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig3-figsupp3-data1-v2.zip
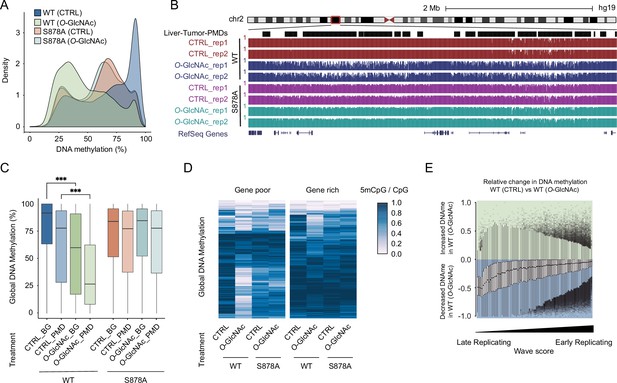
High glucose leads to loss of DNA methylation at cancer-specific partially methylated domains (PMDs).
(A) Density plot of DNA methylation for DNMT1-WT and DNMT1-S878A cells with either low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc). (B) Genome browser screenshot of DNA methylation for DNMT1-WT and DNMT1-S878A cells and low or high glucose along with liver tumor PMDs from Li et al., 2016. (C) Boxplots of DNA methylation at PMDs or general genomic background (BG) for each DNMT1-WT and DNMT1-S878A treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). (D) Heatmap representation of global DNA methylation for DNMT1-WT and DNMT1-S878A cells under low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc) at gene-poor and gene-rich regions. (E) Methylation changes from O-GlcNAcylation of DNMT1 by wave score for replication timing (Hansen et al., 2010; Thurman et al., 2007). ***p<0.0001 by Wilcoxon signed-rank test (C).
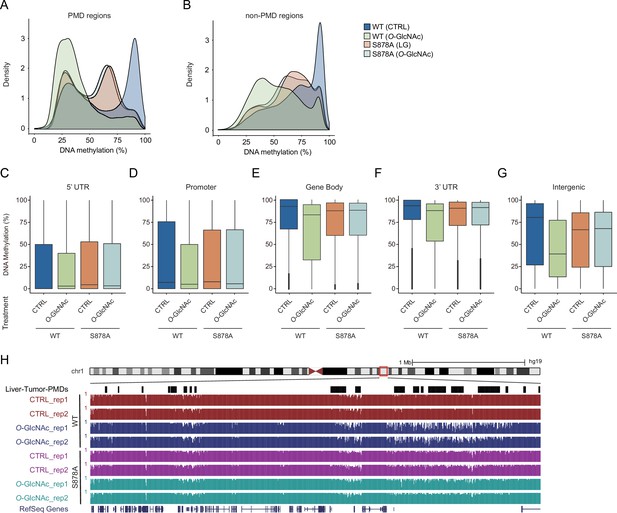
DNA loss of methylation by increased global O-GlcNAcylation decreases.
Density plot of DNA methylation for DNMT1-WT and DNMT1-S878A cells with either low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc). (A) Partially methylated domain (PMD) regions, (B) non-PMD regions. (C–G) Bar graphs represent percentage of global DNA methylation of wild type and DNMT1 mutants (DNMT1-WT or DNMT1-S878A) that treated low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). (C) 5’UTR, (D) promoter, (E) gene body, (F) 3’UTR, and (G) intergenic regions. (H) Genome browser screenshot of DNA methylation data at a differentially methylated region by glucose concentration.
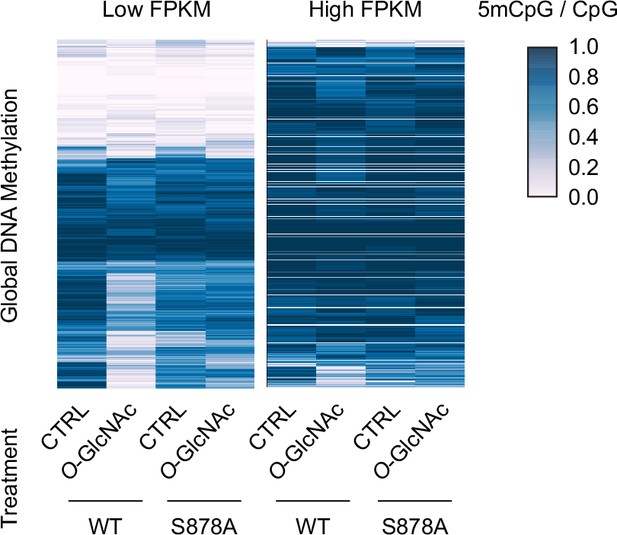
Global DNA methylation of wild type and DNMT1 mutants between low Fragments Per Kilobase of transcript per Million mapped reads (FPKM) regions and high FPKM regions (DNMT1-WT or DNMT1-S878A) that treated low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) were determined by Nanopolish call methylation.
These are defined 'low FPKM' as containing <25% of FPKM regions per Mb window and 'high FPKM' as containing >75% of FPKM regions per Mb window.
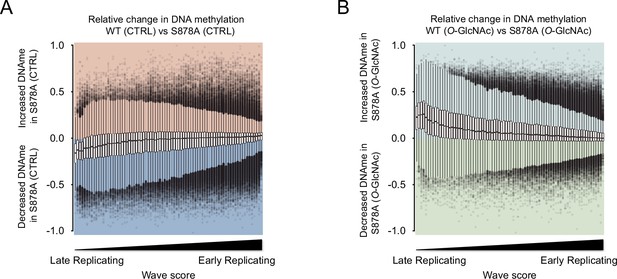
Methylation changes from O-GlcNAcylation of DNMT1 in DNMT1-S878A mutant.
The loss of methylation was not observed in S878A mutant cells. (A, B) The distribution of each DNA methylation was divided by DNA replication timing.
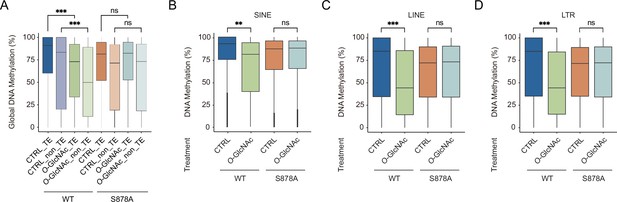
DNA loss of methylation by increased global O-GlcNAcylation decreases around the transposable element (TE) regions.
(A) Boxplot represents the levels of DNA methylation on the TE regions or non-TE regions of each Myc-DNMT1-overexpressed mutants (DNMT1-WT or DNMT1-S878A) were treated with low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc). (B–D) Bar graphs represent percentage of global DNA methylation of wild type and DNMT1 mutants (DNMT1-WT or DNMT1-S878A) that treated low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). (B) SINE, (C) LINE, and (D) LTR regions. **p<0.0005; ***p<0.0001 by Wilcoxon signed-rank test (A–D); ns, not significant.
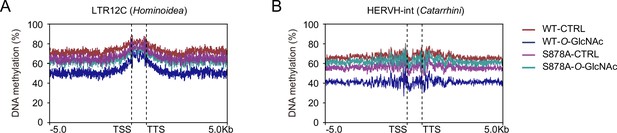
Loss of DNA methylation by increased global O-GlcNAcylation decreases.
Evolutionarily recent TEs are more likely to lose methylation than older elements in a variety of systems. Methylation percentages are shown around (A) LTR12C and (B) HERVH-int regions. The Myc-DNMT1 overexpressed mutants (DNMT1-WT or DNMT1-S878A) were treated low (5mM, CTRL) or high glucose/TMG (25mM, O-GlcNAc).
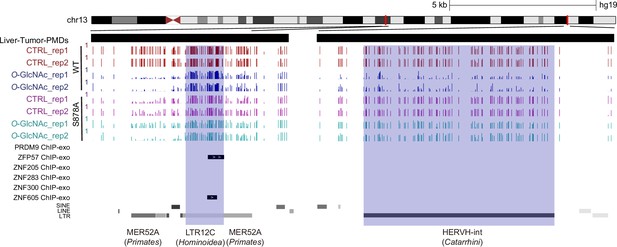
ZFP57 and ZNF605 demonstrate binding to a significant number of LTR12C elements present in liver cancer partially methylated domains (PMDs).
Genome browser screenshot of DNA methylation data LTR12C elements (blue) that demonstrate binding with ZFP57 and ZNF605 by glucose concentration.
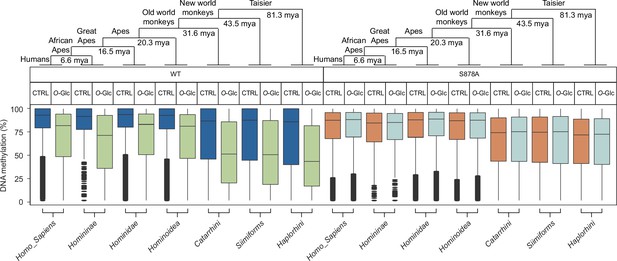
Evolutionarily recent elements are less likely to lose methylation induced by O-GlcNAcylation of DNMT1.
The evolutionary distance between each group is from Perelman et al., 2011. Boxplot represents the DNA methylation by clades of the human genome (Homo sapiens to Haplorhini). CTRL, control; O-Glc, O-GlcNAc.
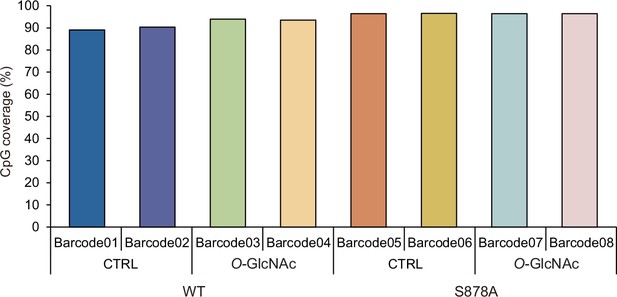
Only loci with >5× coverage were retained for analysis, comprising 90% of CpGs in the genome.
Shown are overall CpGs sites that detected with >5× coverage DNA methylation analysis using Nanopore technology PromethION sequencer. Each condition is biologically replicated.
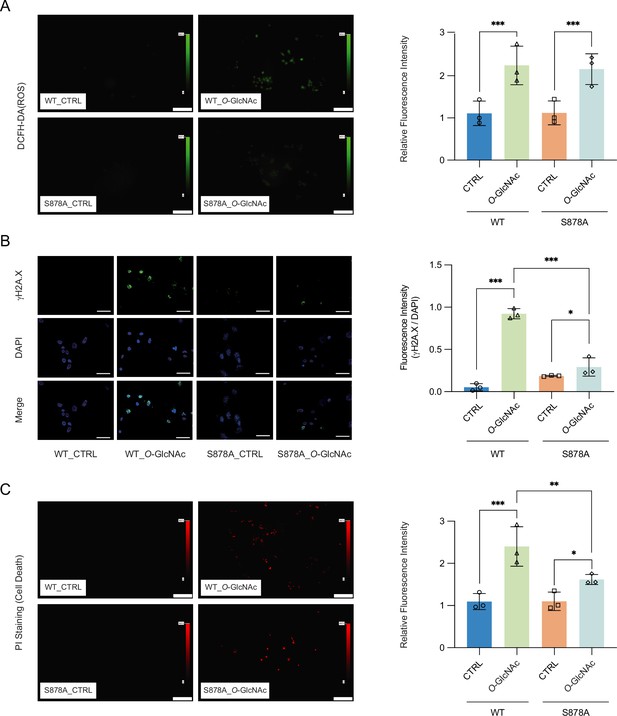
High glucose-induced reactive oxygen species (ROS) and DNA damage cause apoptotic cell death in DNMT1-WT cells.
(A) Quantitative fluorescence image of ROS in DNMT1-WT and DNMT1-S878A cells with either low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc). (B) Quantitative fluorescence image of γ-H2A.X in DNMT1-WT and DNMT1-S878A cells treated with low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). (C) Quantitative fluorescence image of cell death in propidium iodide staining of DNMT1-WT and DNMT1-S878A cells under low (5 mM, CTRL) or high glucose/TMG (25 mM, O-GlcNAc). *p<0.001; **p<0.0005; ***p<0.0001 by Student’s t-test (A–C); data are represented as mean ± SD from three replicates of each sample.
-
Figure 5—source data 1
Raw fluorescence image files of Figure 5A–C.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig5-data1-v2.zip
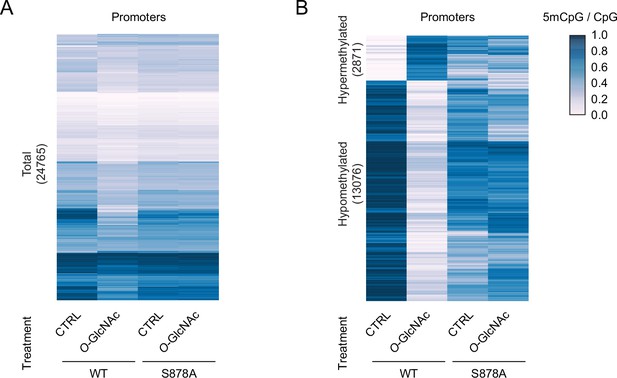
Heatmap representation of promoter DNA methylation for DNMT1-WT and DNMT1-S878A cells under low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) at gene-poor and gene-rich regions.
(A) Whole promoters and (B) differentially methylated promoters.
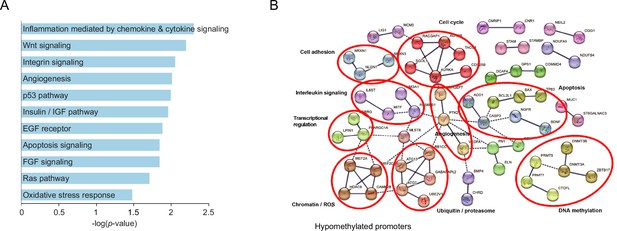
DNA loss of methylation within promoter region by increased global O-GlcNAcylation impact different gene pathways.
(A) Gene Ontology of the top 11 pathways of hypomethylated promoter DNA by high glucose treatment. (B) Gene interaction map of hypomethylated promoter DNA of DNMT1-WT by high glucose treatment.
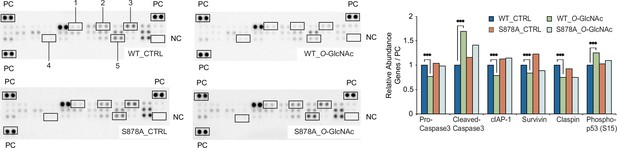
Quantitative analysis of human apoptosis related proteins in DNMT1-WT and DNMT1-S878A with either low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc) using Proteome profiler (n = 3).
PC, positive control; NC, negative control; 1. cleaved-caspase3; 2, cIAP1; 3, claspin; 4, phospho-p53(S15); 5, survivin. ***p<0.0001 by Student’s t-test; Data are represented as mean ± SD from three replicates of each sample.
-
Figure 5—figure supplement 3—source data 1
Uncropped blot files of Figure 5—figure supplement 3.
- https://cdn.elifesciences.org/articles/85595/elife-85595-fig5-figsupp3-data1-v2.zip
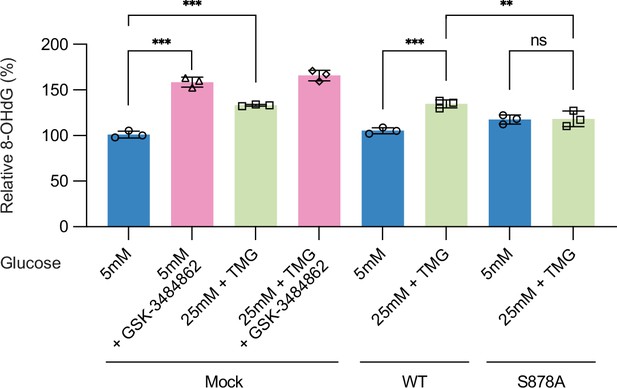
Each Hep3B (Mock) and Myc-DNMT1-overexpressed mutant (DNMT1-WT or DNMT1-S878A) was treated with low (5 mM, CTRL) or high glucose/Thiamet-G (TMG) (25 mM, O-GlcNAc), 5-aza, or GSK-3484862 (negative control).
Shown are absorbance of 8-OHdG performed with DNA damage quantification kit (n = 3). **p<0.0005; ***p<0.0001 by Student’s t-test; ns, not significant; data are represented as mean ± SD from three replicates of each sample.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | DNMT1 | HUGO Gene Nomenclature Committee | HGNC:2976 | - |
| Cell line (H. sapiens) | Hep 3B2.1–7 | ATCC | HB-8064 | - |
| Cell line (H. sapiens) | Hep G2 | ATCC | HB-8065 | - |
| Transfected construct (H. sapiens) | pcDNA3/Myc-DNMT1 | Addgene | Plasmid #36939 | |
| Antibody | Anti-beta-actin (D6A8) (rabbit monoclonal) | Cell Signaling Technology | Cat# 8457 | WB (1:1000) |
| Antibody | Anti-alpha-tubulin (11H10) (rabbit monoclonal) | Cell Signaling Technology | Cat# 2125 | WB (1:1000) |
| Antibody | Anti-DNMT1 (60B1220.1) (mouse monoclonal) | Novus Biologicals | Cat# NB100-56519 | IP (1:250) WB (1:1000) |
| Antibody | Anti-DNMT1 (H-12) (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-271729 | WB (1:1000) |
| Antibody | Anti-gamma H2A.X (rabbit polyclonal) | Abcam | Cat# ab11174 | IF (1:1000) |
| Antibody | Anti-GAPDH (rabbit monoclonal) | Abcam | Cat# ab181602 | WB (1:1000) |
| Antibody | Anti-H3 (rabbit polyclonal) | Abcam | Cat# ab1791 | WB (1:1000) |
| Antibody | Anti-Myc [Myc.A7] (mouse monoclonal) | Abcam | Cat# ab18185 | IP (1:250) WB (1:1000) |
| Antibody | Anti-O-GlcNAc (RL2) (mouse monoclonal) | Abcam | Cat# ab2739 | WB (1:1000) |
| Antibody | Aoat anti-rabbit IgG H&L (HRP) (goat polyclonal) | Abcam | Cat# ab6721 | WB (1:5000) |
| Antibody | Goat anti-mouse IgG H&L (HRP) (goat polyclonal) | Abcam | Cat# ab6789 | WB (1:5000) |
| Antibody | Goat anti-mouse IgG (H+L), Alexa 488 (goat polyclonal) | Invitrogen | Cat# A32723 | WB (1:1000) |
| Sequence-based reagent | DNMT1-T158A | This paper | PCR primers | agccccaggatt CGA aggaaaagcacc |
| Sequence-based reagent | DNMT1-T158A | This paper | PCR primers | ggtgcttttcct TCG aatcctggggct |
| Sequence-based reagent | DNMT1-T616A | This paper | PCR primers | gacaggggaccc GCG aaagccaccacc |
| Sequence-based reagent | DNMT1-T616A | This paper | PCR primers | ggtggtggcttt CGC gggtcccctgtc |
| Sequence-based reagent | DNMT1-S878A | This paper | PCR primers | gcgagattcgag GAG cctccaaaaacc |
| Sequence-based reagent | DNMT1-S878A | This paper | PCR primers | ggtttttggagg CTC ctcgaatctcgc |
| Sequence-based reagent | DNMT1-S878D | This paper | PCR primers | gcgagattcgag GAC cctccaaaaacc |
| Sequence-based reagent | DNMT1-S878D | This paper | PCR primers | ggtttttggagg GTC ctcgaatctcgc |
| Sequence-based reagent | DNMT1-T882A | This paper | PCR primers | tcccctccaaaa GCC cagccaacagag |
| Sequence-based reagent | DNMT1-T882A | This paper | PCR primers | ctctgttggctg GGC ttttggagggga |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis kit | NEB | Cat# E0554S | - |
| Commercial assay or kit | EpiQuik DNMT Activity/Inhibition ELISA Easy Kit | EpiGentek | Cat# P-3139 | - |
| Commercial assay or kit | Global DNA methylation LINE-1 | Active Motif | Cat# 55017 | - |
| Commercial assay or kit | EpiQuik 8-OHdG DNA Damage Quantification Direct kit | EpiGentek | Cat# P-6003 | - |
| Chemical compound, drug | OSMI-4 | Selleck Chem | Cat# S8910 | - |
| Chemical compound, drug | Thiamet-G (TMG) | Cayman Chemical | Cat# 13237 | - |
| Software, algorithm | bedGraphToBigWig | Kent et al., 2010 | - | - |
| Software, algorithm | Clustal Omega | Sievers et al., 2011 | - | Version 1.2.4 |
| Software, algorithm | GraphPad Prism 9 | GraphPad | - | Version: 9.3.1 |
| Software, algorithm | Minimap2 | Li and Birol, 2018 | RRID:SCR_018550 | Version: 2.17 |
| Software, algorithm | Nanopolish | Loman et al., 2015 | RRID:SCR_016157 | Version: 0.11.1 |
| Software, algorithm | Python | Python Core Team | Version: 3.8.2 | |
| Software, algorithm | R | R Core Team | - | Version: 3.4.3 |
| Software, algorithm | Samtools | Lister et al., 2009 | RRID:SCR_002105 | Version: 1.10 |
| Other | UniProt | The UniProt Consortium | - | Database of protein information (https://www.uniprot.org/) |
| Other | DAPI stain | Invitrogen | D1306 | 1 µg/ml; marker for nuclear DNA |
Additional files
-
Supplementary file 1
Prediction of O-GlcNAcylated sites within DNMT1 using OGTSite.
- https://cdn.elifesciences.org/articles/85595/elife-85595-supp1-v2.docx
-
Supplementary file 2
List of total identified proteins.
- https://cdn.elifesciences.org/articles/85595/elife-85595-supp2-v2.docx
-
Supplementary file 3
List of post-translational modification (PTM) sites of human DNMT1.
- https://cdn.elifesciences.org/articles/85595/elife-85595-supp3-v2.docx
-
Supplementary file 4
List of deposited data in this study.
- https://cdn.elifesciences.org/articles/85595/elife-85595-supp4-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85595/elife-85595-mdarchecklist1-v2.docx