Interplay between acetylation and ubiquitination of imitation switch chromatin remodeler Isw1 confers multidrug resistance in Cryptococcus neoformans
Figures
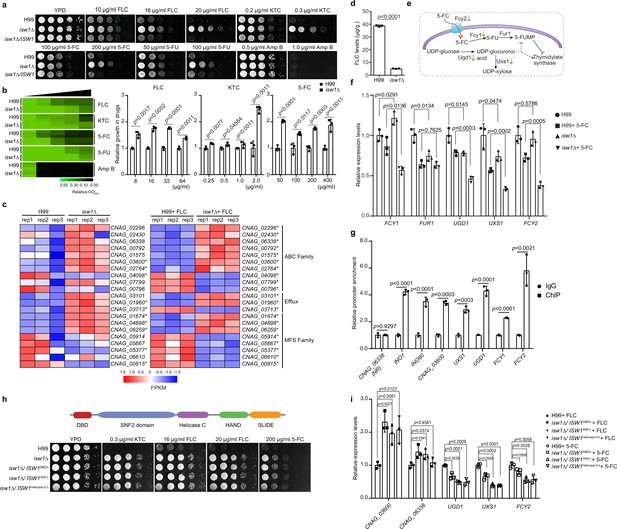
Isw1 represses the expression of drug-resistance genes.
(a) Spotting assays of ISW1 mutant strains. Wild-type H99, isw1Δ, and ISW1 complementation strains were spotted onto YPD agar supplemented with indicated concentrations of antifungal agents. Plates were incubated at 30°C for 3 days. (b) Drug inhibitory tests. The H99 and isw1Δ (n = 3 each) strains were tested to determine the drug inhibition of several antifungal agents. Twofold dilutions of fluconazole (FLC) from 0 to 64 µg/ml, ketoconazole (KTC) from 0 to 2 µg/ml, or 5-fluorocytosine (5-FC) from 0 to 400 µg/ml were added to YPD medium. After 24 hr at 30°C, absorbance at 600 nm was used to measure growth. The optical densities of duplicate measurements were averaged and normalized relative to the control without FLC. Quantitative data were depicted in color (see color bar) and bar plots. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (c) Transcriptome analysis of isw1Δ. Samples of RNA were isolated from H99 and isw1Δ cells (n = 3 each) supplemented with or without 10 μg/ml FLC. Transcriptome analysis was performed, and a heat map of the expressions of drug-resistance genes was generated. Statistical significant genes are labeled with asterisks. (d) Intracellular concentration of FLC. H99 and isw1Δ cells (n = 3 each) were incubated in the presence of 40 μg/ml FLC at 30°C for 5 hr. Cells were then washed and weighed, and the intracellular FLC was quantified using high-performance liquid chromatography. A two-tailed unpaired t-test was used. Data are expressed as mean ± SD. (e) Scheme of the mechanism for 5-FC and 5-fluorouracil (5-FU) resistance. The red arrow indicates the entry of 5-FC via Fcy2. Green arrows indicate the downregulation of gene expression in response to 5-FC in the isw1Δ strain. (f) Analyses of 5-FC-resistance genes using quantitative reverse transcription PCR(qRT-PCR). Indicated strains (n = 3 each) were grown with or without 400 μg/ml 5-FC, then qRT-PCR was performed to determine gene expressions of FCY1, FUR1, UGD1, UXS1, and FCY2. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (g) Chromatin immunoprecipitation (ChIP)-PCR analysis of Isw1-FLAG. ISW1-FLAG cells (n = 3) were incubated without drug treatment at 30°C for 5 hr. ChIP-PCRs were performed in the ISW1-FLAG strain (n = 3 each). Isw1 target genes, INO1 and INO80, were amplified and used as positive controls. CNAG_06338 were used as a negative control. Potential target genes, CNAG_03600, UXS1, UGD1, FCY1, and FCY2 were tested. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (h) Truncation analysis of cryptococcal Isw1. Truncated Isw1s were cloned and transformed into the isw1Δ strain to generate the DNA-binding domain (DBD) truncation (isw1Δ/ISW1DBDΔ), SNF truncation (isw1Δ/ISW1SNFΔ) and helicase C truncation (isw1Δ/ISW1Helicase-CΔ) strains, which were spotted onto YPD agar plates supplemented with indicated antifungal drugs. (i) qRT-PCR analysis in truncated strains supplemented with 5-FC or FLC. Indicated strains were treated with 5-FC or FLC. RNA samples were isolated. qRT-PCRs were performed using oligos from UGD1, UXS1, FCY2, CNAG_03600, and CNAG_06338.
-
Figure 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig1-data1-v2.zip

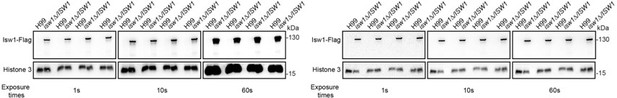
Construction of the ISW1-FLAG complementation strain.
(a) qRT-PCR of ISW1. Samples of RNA were isolated from H99 cells treated with fluconazole (FLC) or 5-fluorocytosine (5-FC). qRT-PCR was performed on each to confirm the gene expression of ISW1. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (b) Immunoblotting analysis of the ISW1 complementation strain. The complementation strain was constructed, and an immunoblotting assay was performed to confirm the expression of the Isw1-Flag protein. (c) Genomic copy number assays. Genomic DNA was isolated from the wild-type and complementation strains, then qRT-PCR was performed on each to confirm the gene copy number of ISW1. Oligos of actin were used as a control (Supplementary file 3). Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (d) Analyses of the ISW1 complementation strain using qRT-PCR. Samples of RNA were isolated from the wild-type and complementation strains, then qRT-PCR was performed on each to confirm the gene expression of ISW1. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD.
-
Figure 1—figure supplement 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig1-figsupp1-data1-v2.zip
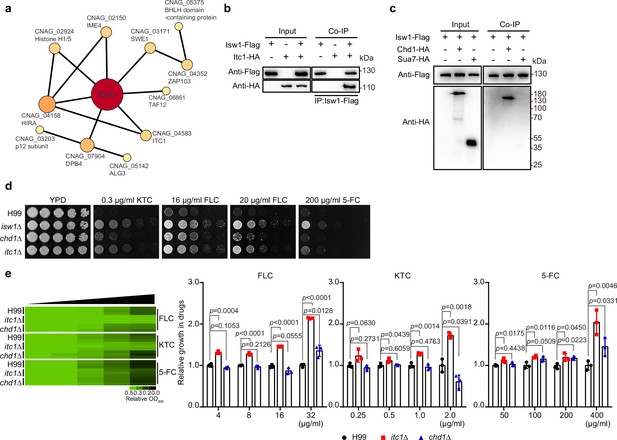
Characterization of Isw1-interacting proteins.
(a) Mass spectrometry analysis of Isw1 regulating network. Protein co-immunoprecipitation (co-IP) assays were carried out, followed by mass spectrometry analysis. Isw1-interacting proteins were identified, and interacting network was generated using StringDB. (b) Isw1 and Itc1 co-IP analysis. Proteins were isolated from strains expressing Isw1-FLAG or Itc1-HA or both, and co-IP followed by immunoblotting assays were performed. Antibodies against FLAG and HA epitope tags were used. (c) Isw1, Sua1, and Chd1 co-IP analysis. Proteins were isolated from strains expressing Isw1-FLAG, Isw1-FLAG/Chd1-HA, or Isw1-FLAG/Sua7-HA, and co-IP followed by immunoblotting assays were performed. Antibodies against FLAG and HA epitope tags were used. (d) Spotting analysis of chd1Δ and itc1Δ strains. Indicated strains were spotted onto YPD agar plates supplemented with antifungal agents. (e) Drug inhibitory tests of chd1Δ and itc1Δ strains. The H99, chd1Δ, and itc1Δ strains (n = 3 each) were tested to determine inhibitory effects of several antifungal agents. The experiments were performed as described in Figure 1b. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD).
-
Figure 2—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig2-data1-v2.zip
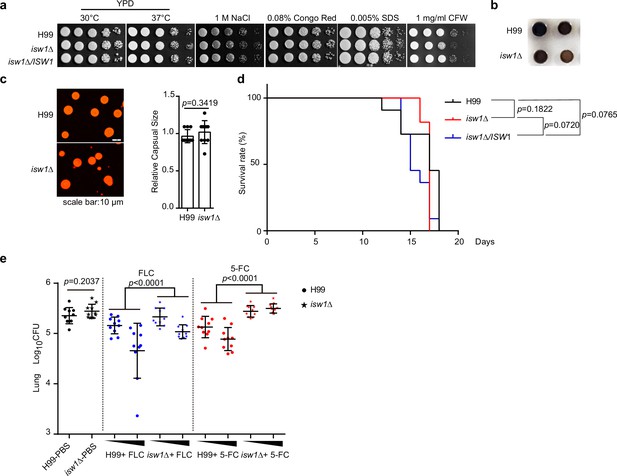
Isw1 plays a critical role in drug resistance during pulmonary infection.
(a) Spotting analysis of isw1Δ in stress conditions. H99, isw1Δ, and ISW1 complementation strains were spotted onto YPD agar plates supplemented with indicated chemicals. (b) Melanin formation of the isw1Δ strain. (c) Capsule formation and quantification of the isw1Δ strain. H99 and isw1Δ cells were induced for capsule structure. Capsular sizes were quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (d) Animal survival analysis and the Kaplan–Meier survival curves of wild-type and isw1Δ. Significance was determined using a log-rank (Mantel–Cox) test. (e) Colony-forming unit (CFU) analysis of the isw1Δ strain. Mice (n = 10 each) were infected with H99 or isw1Δ cells. At 7-day post infection, animals were treated with phosphate-buffered saline (PBS), or 5 or 45 mg/kg of fluconazole (FLC), or 100 or 200 mg/kg 5-fluorocytosine (5-FC). Animals were treated with drugs in a 24-hr interval on a daily basis. At 14-day post infection, lung tissues were removed and homogenized. CFUs were performed. Data are expressed as mean ± standard deviation (SD). Two-tailed unpaired t-tests were used for H99-PBS and isw1Δ-PBS group. Two-way analyses of variance (ANOVAs) were used for drug-treated groups.
-
Figure 3—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig3-data1-v2.zip
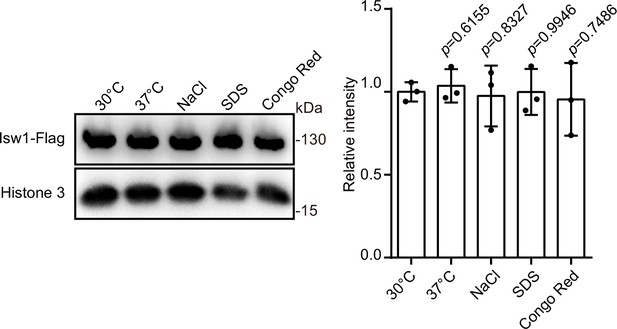
Isw1 is not responsive to other environmental stresses.
Immunoblotting analysis and quantification of Isw1-FLAG in stress conditions. Samples (n = 3 each) of protein were isolated from ISW-FLAG cells incubated in the indicated stresses conditions, and an immunoblotting assay was performed to confirm the expression of the Isw1-Flag protein. Data are expressed as mean ± standard deviation (SD).
-
Figure 3—figure supplement 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig3-figsupp1-data1-v2.zip
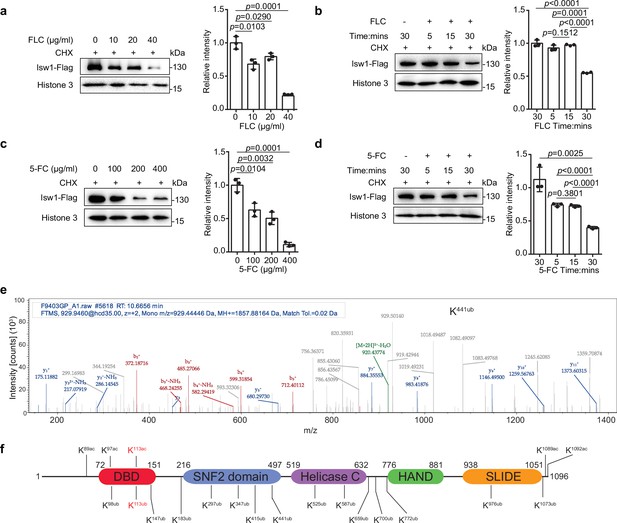
Isw1 is an acetylated and ubiquitinated protein.
(a) Immunoblotting analysis. The ISW1-FLAG strain was preincubated with 200 μM cycloheximide (CHX) for 1 hr followed by exposure to various concentrations of fluconazole (FLC) for 0.5 hr. Anti-Flag and anti-histone 3 antibodies were used. Three biological replicates were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (b) Immunoblotting analysis. The ISW1-FLAG strain was preincubated with 200 μM cycloheximide (CHX) for 1 hr followed by exposure to 40 μg/ml FLC for 5, 15, or 30 min. Cells not exposed to FLC but held for 30 min were used as a negative control. Anti-Flag and anti-histone 3 antibodies were used. Three biological replicates were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (c) Immunoblotting analysis. Testing and data treatment were exactly as described for Figure 2a with the exception that 5-fluorocytosine (5-FC) was used as the antifungal agent. (d) Immunoblotting analysis. Testing and data treatment were exactly as described in Figure 2b, except that 5-FC was used as the antifungal agent. (e) Ubiquitin analysis of Isw1 via mass spectrometry. The Isw1-Flag proteins were pulled down and analyzed for ubiquitination. Results for Isw1K441Ub are shown. (f) Schematic of Isw1 showing acetylation (Li et al., 2019) and ubiquitination sites.
-
Figure 4—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig4-data1-v2.zip
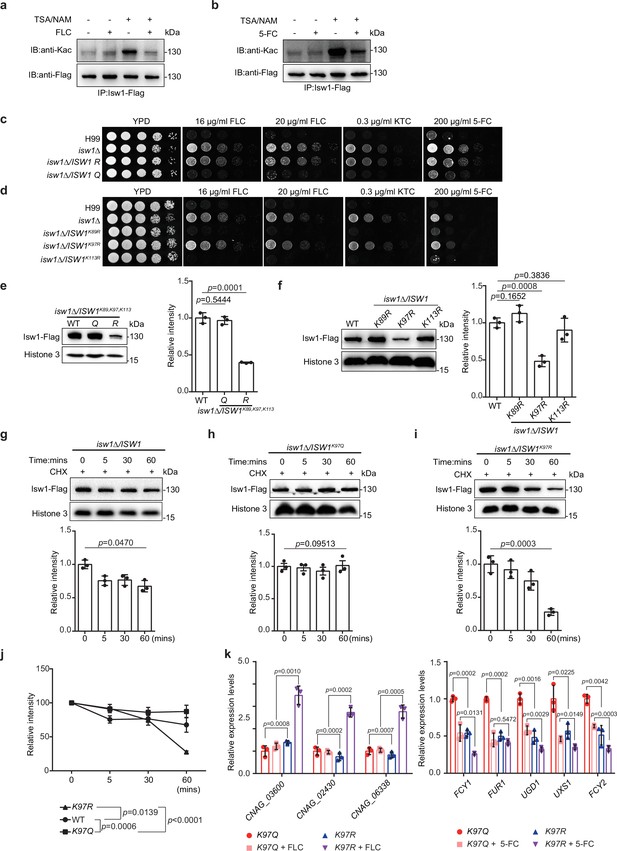
The acetylation status of Isw1K97 (Isw1K97ac) is essential in Isw1 protein stability.
(a) Acetylation analysis of Isw1. Cells were treated with 3 μM trichostatin A (TSA), 20 mM nicotinamide (NAM), and fluconazole (FLC). The Isw1-Flag proteins were pulled down, and immunoblotting assays were performed using anti-Kac and anti-Flag antibodies. (b) Acetylation analysis of Isw1. Cells were treated with TSA, NAM, and 5-fluorocytosine (5-FC). The Isw1-Flag proteins were pulled down, and immunoblotting assays were performed using anti-Kac and anti-Flag antibodies. (c) Spotting assays of ISW1 mutants. The ISW1K89R, K97R, K113R and ISW1K89Q, K97Q, K113Q strains were tested for drug resistance. (d) Spotting assays of ISW1 mutants. The ISW1K89R, ISW1K97R, and ISW1K113R strains were tested for drug resistance. (e) Immunoblotting assays of ISW1 mutants. The wild-type, ISW1K89R, K97R, K113R, and ISW1K89Q, K97Q, K113Q strains were tested for Isw1 levels. Three biological replicates were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (f) Immunoblotting assays of ISW1 mutants. The wild-type, ISW1K89R, ISW1K97R, and ISW1K113R strains were tested for Isw1 levels. Three biological replicates were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (g) Immunoblotting assay of Isw1. The wild-type strain was preincubated with 200 μM cycloheximide for 1 hr. Proteins were isolated at indicated time points. Three biological replicates of immunoblotting were performed, and results were used for quantification. One-way analysis of variance (ANOVA) was used. (h) Immunoblotting assay of Isw1K97Q. The analysis was performed as described in Figure 3g. (i) Immunoblotting assay of Isw1K97R. The analysis was performed as described in Figure 3g. (j) Comparisons of assay results to determine Isw1 stability. The relative intensities from the results shown in Figure 3g–i were plotted. Two-way ANOVA was used. Data are expressed as mean ± SD. (k) Analyses of drug-resistance genes using qRT-PCR. Samples of RNA (n = 3) were isolated from ISW1K97Q and ISW1K97R treated with FLC or 5-FC. Representative drug-resistance genes were quantified using qRT-PCR. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD.
-
Figure 5—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig5-data1-v2.zip

Screening important acetylation sites of Isw1.
(a) Genomic copy number assays. Genomic DNA was isolated from the indicated ISW1 mutants, then qRT-PCR was performed on each to confirm the gene copy number of ISW1. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (b) Analyses of ISW1 mutants using qRT-PCR. Samples of RNA were isolated from the wild-type and indicated mutants, then qRT-PCR was performed on each to confirm the gene expressions of wild-type and mutated ISW1. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (c) Spotting assays of ISW1 double-R mutants. The wild-type and indicated ISW1 mutants were spotted onto YPD agar either supplemented with an antifungal agent or left blank. (d) Spotting assays of ISW1 single-Q mutants. The wild-type and indicated ISW1 mutants were spotted onto YPD agar either supplemented with an antifungal agent or left blank. (e) Immunoblotting analyses of ISW1 single-Q mutants. Protein samples were isolated from the indicated strains, and immunoblotting assays were performed.
-
Figure 5—figure supplement 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig5-figsupp1-data1-v2.zip
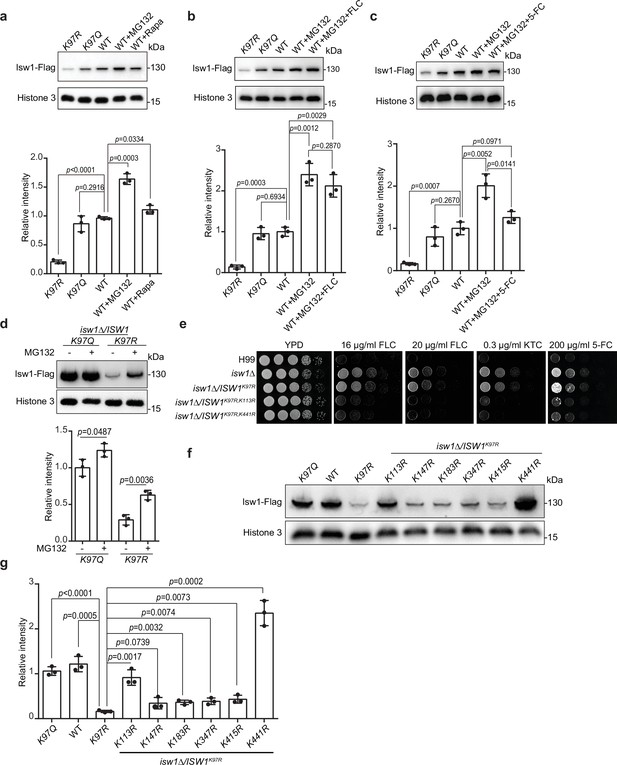
Isw1K97ac is critical for Isw1 ubiquitin–proteasome degradation.
(a) Immunoblotting assays of Isw1-Flag. Proteins Isw1K97Q, Isw1K97R, and Isw1 were tested, where the wild-type stain was incubated with 200 μM MG132 and 5 nM rapamycin for 10 hr. Three biological replicates of the assays were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (b) Immunoblotting assays of Isw1-Flag. Testing and data treatment were exactly as described in Figure 4a, except that the wild-type sample was treated with 200 μM of MG132 and 40 μg/ml fluconazole (FLC). (c) Immunoblotting assays of Isw1-Flag. Testing and data treatment were exactly as described in Figure 4a, except that the wild-type sample was treated with 200 μM MG132 and 400 μg/ml 5-fluorocytosine (5-FC). (d) Immunoblotting assays of Isw1K97Q and Isw1K97R. Proteins were either treated with MG132 or not before testing. Three biological replicates of the assays were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (e) Spotting assays of ISW1 acetylation and ubiquitination mutants. Indicated strains were spotted onto YPD agar either supplemented with an antifungal agent or left blank. (f) Immunoblotting assays of ISW1 acetylation and ubiquitination mutants. Protein samples were isolated from the indicated ISW1 mutants. Immunoblotting assays were performed. (g) Quantification of immunoblotting results. The immunoblotting assays described for Figure 3f were performed using three independent samples, and the results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD.
-
Figure 6—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig6-data1-v2.zip
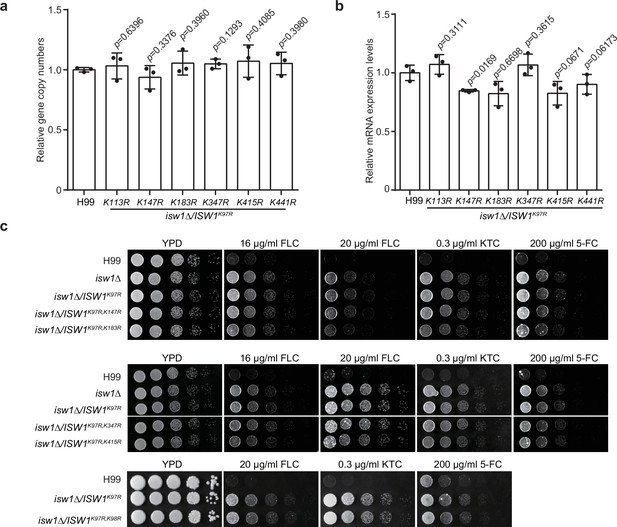
Screening important ubiquitination sites of Isw1.
(a) Genomic copy number assays. Genomic DNA was isolated from indicated ISW1 mutants, then qRT-PCR was performed on each to confirm the gene copy number of ISW1. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (b) Analyses of ISW1 ubiquitination mutants using qRT-PCR. Samples of RNA were isolated from wild-type and indicated mutants, and qRT-PCR was performed on each to confirm the expressions of wild-type and mutated ISW1 genes. Oligos of actin were used as a control. Three independent assays were performed and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (c) Spotting assays of ISW1 ubiquitination site mutants. The wild-type and indicated ISW1 mutants were spotted onto YPD agar either supplemented with an antifungal agent or left blank. Plates were incubated at 30°C for 3 days.
-
Figure 6—figure supplement 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig6-figsupp1-data1-v2.zip
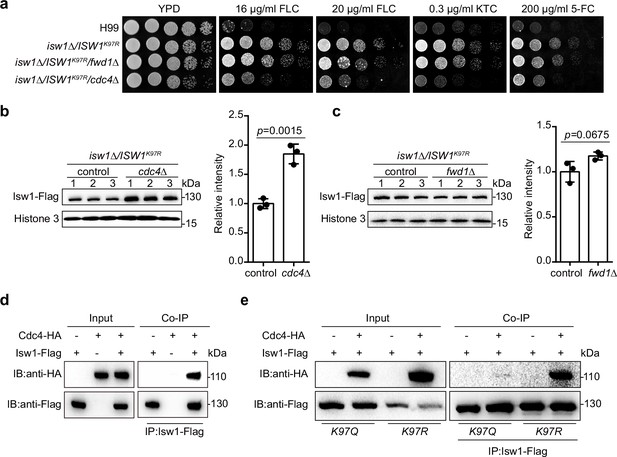
IswK97ac blocks the binding of Isw1 to the E3 ligase Cdc4.
(a) Spotting assays of E3 ligase mutants. Indicated strains were spotted onto YPD agar either supplemented with an antifungal agent or left blank. (b) Immunoblotting assays of cdc4Δ. Protein samples were isolated from isw1Δ/ISW1K97R/cdc4Δ and its relevant control strains. Three independent samples were tested and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (c) Immunoblotting assays of fwd1Δ. Protein samples were isolated from isw1Δ/ISW1K97R/fwd1Δ and its relevant control strains. Three independent samples were tested and quantified. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD. (d) Protein co-immunoprecipitation (co-IP) of Cdc4 and Isw1. Protein samples were isolated from the strain expressing Cdc4-HA and Isw1-Flag, and co-IP was performed. (e) Protein co-IP of Cdc4 and Isw1 K97 mutant proteins. Protein samples were isolated from the strain co-expressing Cdc4-HA and Isw1K97R-Flag and the strain co-expressing Cdc4-HA and Isw1K97Q-Flag. Co-IP was performed for each.
-
Figure 7—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig7-data1-v2.zip

Identification of the E3 ligase for Isw1.
(a) Spotting assays of E3 ligase knockout strains. Wild-type and indicated E3 ligase knockout strains were spotted onto YPD agar either supplemented with an antifungal agent or left blank. Plates were incubated at 30°C for 3 days. (b) Spotting assays of cdc4Δ. Wild-type and indicated knockout strains were spotted onto YPD agar either supplemented with an antifungal agent or left blank. Plates were incubated at 30°C for 3 days.
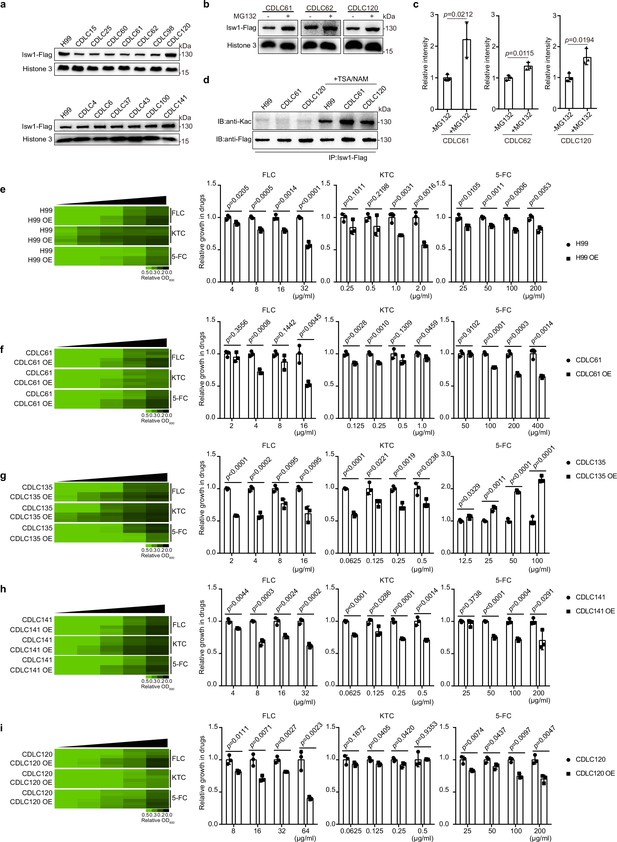
Clinical C. neoformans isolates show Isw1-mediated drug-resistance phenotypes.
(a) Immunoblotting assays of Isw1-Flag from clinical isolates. Protein samples were isolated from H99 and clinical isolates expressing Isw1-Flag. Immunoblotting analyses were performed. (b) Immunoblotting analyses of Isw1 levels under MG132 treatment. Cells were treated with MG132, then immunoblotting assays were performed on Isw1-Flag using anti-Flag antibodies. (c) Three independent repetitions from Figure 8b were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (d) Immunoblotting analyses of Isw1 acetylation in clinical strains. Cells were treated with trichostatin A and nicotinamide, then Isw1-Flag was immunoprecipitated. Acetylation levels were examined using anti-Kac antibodies. (e–i) Drug inhibitory analyses of Isw1-overexpressing clinical strains. Clinical strains harboring integrative overexpressing plasmid of ISW1K97Q were tested for drug resistance, and minimum inhibitory concentrations (MICs) were determined. OE indicates strains with overexpressed ISW1K97Q. The experiments were performed as described in Figure 1b. Quantitative data were depicted in color (see color bar) and bar plots. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD.
-
Figure 8—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig8-data1-v2.zip
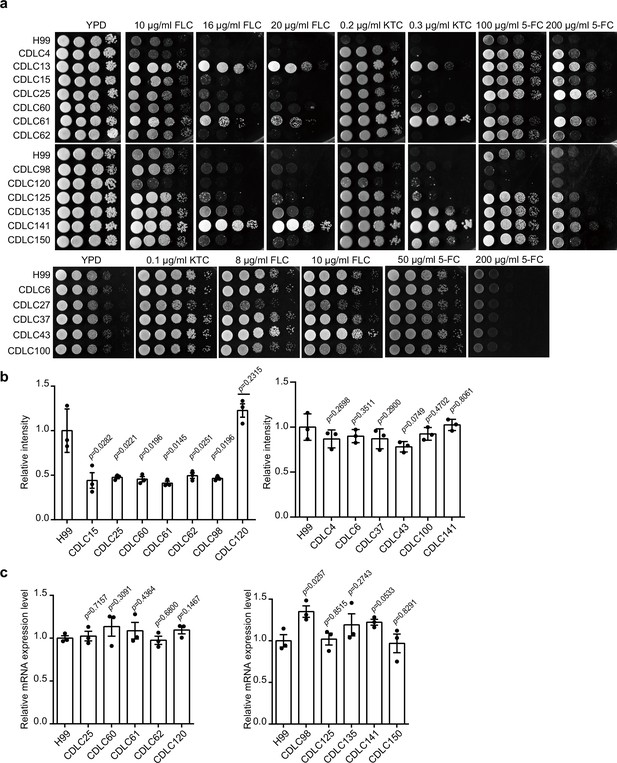
Drug-resistance analysis of clinical isolates.
(a) Spotting assays of C. neoformans clinical strains. Clinical strains were spotted onto YPD agar either supplemented with an antifungal agent or left blank. Results after 3 days of incubation at 30°C are shown. (b) Three independent repetitions from Figure 8a were performed, and results were used for quantification. Two-tailed unpaired t-tests were used. Data are expressed as mean ± standard deviation (SD). (c) Analyses of ISW1 in clinical isolates using qRT-PCR. Samples of RNA were isolated from H99 and clinical strains, then qRT-PCR performed on each followed by quantification of ISW1. Three biological replicates were performed. Two-tailed unpaired t-tests were used. Data are expressed as mean ± SD.
-
Figure 8—figure supplement 1—source data 1
The source data comprises unprocessed data.
- https://cdn.elifesciences.org/articles/85728/elife-85728-fig8-figsupp1-data1-v2.zip
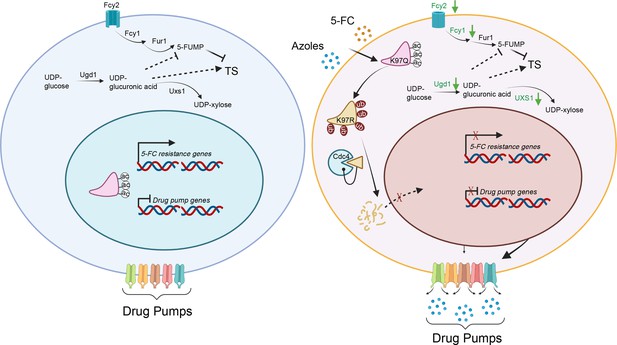
A model of the mechanism of Isw1 posttranslational modification (PTM) interaction in C. neoformans drug resistance.
In a drug-free environment, acetylated Isw1 regulates 5-fluorocytosine (5-FC)-resistance gene expression and represses drug pump gene expression. Azoles and 5-FC trigger the deacetylation process at the K97 residue of Isw1, initiating the ubiquitin-mediated proteasomal degradation of Isw1 through the E3 ligase Cdc4. The decrease in Isw1 protein level results in the stimulation of drug pump gene expression and the inhibition of 5-FC-resistance genes.
Additional files
-
Supplementary file 1
Strains used in this study.
- https://cdn.elifesciences.org/articles/85728/elife-85728-supp1-v2.xlsx
-
Supplementary file 2
DEGs (differentially expressed genes) in isw1Δ cells treated with or without fluconazole (FLC).
- https://cdn.elifesciences.org/articles/85728/elife-85728-supp2-v2.xlsx
-
Supplementary file 3
Primers used in this study.
- https://cdn.elifesciences.org/articles/85728/elife-85728-supp3-v2.xlsx
-
Supplementary file 4
Co-immunoprecipitation (Co-IP) mass spectrometry analysis of Isw1-Flag.
- https://cdn.elifesciences.org/articles/85728/elife-85728-supp4-v2.xlsx
-
Supplementary file 5
Comparison of drug-resistant genes among clinical isolates.
- https://cdn.elifesciences.org/articles/85728/elife-85728-supp5-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85728/elife-85728-mdarchecklist1-v2.docx