The myocardium utilizes a platelet-derived growth factor receptor alpha (Pdgfra)–phosphoinositide 3-kinase (PI3K) signaling cascade to steer toward the midline during zebrafish heart tube formation
Figures
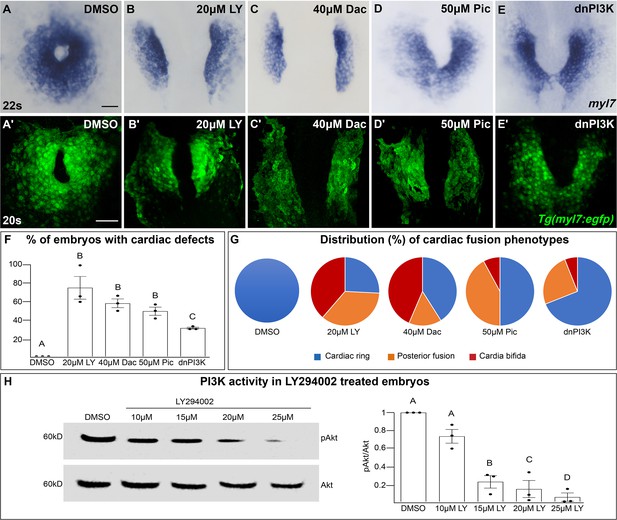
The phosphoinositide 3-kinase (PI3K) pathway is required for cardiac fusion.
Dorsal views, anterior to the top, of the myocardium labeled with myl7 (A–E) at 22 somite stage (s) or Tg(myl7:egfp) (A'–E') at 20s. In contrast to a ring of myocardial cells in DMSO-treated embryos (A, A'), in embryos treated with PI3K inhibitors LY294002 (LY, B, B'), Dactolisib (Dac, C, C'), or Pictilisib (Pic, D, D') at bud stage or injected with dnPI3K mRNA (750 pg) at the one-cell stage (E, E') cardiac fusion fails to occur properly with embryos displaying either cardia bifida (B, C) or fusion only at the posterior end (D, E). Graphs depict the percentage (F) and range (G) of cardiac fusion defects in control and PI3K-inhibited embryos. Dots represent the percent of embryos with cardiac defects per biological replicate. Total embryos analyzed n = 37 (DMSO), 31 (20 μM LY), 39 (40 μM Dac), 38 (50 μM Pic), and 86 (dnPI3K). Blue – cardiac ring/normal; orange – fusion only at posterior end/mild phenotype, red – cardia bifida/severe phenotype. (H) Representative immunoblot and ratiometric analysis of phosphorylated Akt (pAkt) to Akt protein levels in DMSO- and LY-treated embryos reveals a dose-dependent decrease in PI3K activation. Bar graphs indicate mean ± standard error of the mean (SEM), dots indicate pAKT/AKT ratio per biological replicate, normalized to DMSO. Three biological replicates per treatment. One-way analysis of variance (ANOVA) tests – letter changes indicate differences of p < 0.05 (F, H). Scale bars, 40 μm (A–E), 42 μm (A'–E'). Raw data and full p-values included in the source file.
-
Figure 1—source data 1
Statistical source data for Figure 1F, H.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Original immunoblots used in Figure 1H (raw, uncropped) with and without labeling.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-data2-v3.zip

The penetrance and severity of cardiac fusion defects in phosphoinositide 3-kinase (PI3K)-inhibited embryos is dose dependent.
(A–L) myl7 insitus labeling the myocardium at 22s. Incubation with LY (A–C), Dac (D–F), Pic (G–I) from bud stage to 22s or injection with dnPI3K mRNA (J–L) at the one-cell stage results in dose-dependent cardiac fusion defects at 22s. Graphs depict the distribution of cardiac fusion defects in embryos treated with increasing concentrations of LY (M), Dac (N), Pic (O), or dnPI3K mRNA (P). Both the percent of embryos displaying cardiac fusion defects and the severity of those defects are dose dependent. Number of embryos analyzed (n) at the indicated concentrations in (M–P) LY-40, 40, 30, 31, 31; Dac: 38, 34, 39; Pic: 37, 39, 38; dnPI3K mRNA: 73, 52, 61, 57, 52, respectively. Dots indicate the percent of embryos displaying a specific phenotype per incubation. Blue – cardiac ring/normal; orange – fusion only at posterior end/mild, red – cardia bifida/severe. Bar graphs, mean ± standard error. Representative immunoblot and ratiometric analysis of phosphorylated Akt (pAkt) to Akt protein levels in DMSO and Dac (Q), Pic (R), and dnPI3K mRNA (S) treated embryos reveals a dose-dependent decrease in PI3K activation. Bar graphs indicate mean ± standard error, dots indicate pAKT/AKT ratio per biological replicate, normalized to DMSO. At least three biological replicates per treatment. Letter change indicates p < 0.05, one-way analysis of variance (ANOVA). Scale = 60 μm. Raw data with full p-values included in the source file.
-
Figure 1—figure supplement 1—source data 1
Statistical source data for Figure 1—figure supplement 1M–O, P–S.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp1-data1-v3.xlsx
-
Figure 1—figure supplement 1—source data 2
Original immunoblots used in Figure 1—figure supplement 1Q–S (raw, uncropped) with and without labeling.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp1-data2-v3.zip

LY incubation results in trunk extension and somite formation delays.
Lateral brightfield views of 20 hours post-fertilization (hpf) embryos treated with DMSO (A, D) or 20 μM LY (B, E) at bud stage. (C, F) Box-whisker plot depicting the median embryonic length (yellow curved line in A, B) or somite number (yellow dots in D, E) at 20 hpf. Total number of embryos (n) from >3 separate incubations = 40 (DMSO), 40 (20 μM LY) for (C), and 39 (DMSO), 42 (20 μM LY) for (F). Dots = measurements from individual embryos. Two-sample t-test; p-value = 4.527 × 10−4 and 7.624 × 10−5, respectively. (G–H) Dorsal views, anterior to the top, of the myocardium labeled with myl7 at 20 hpf. Embryos treated with DMSO at bud stage show cardiac rings (G) whereas those treated with 20 μM LY show cardia bifida at 20 hpf (H). (I) Graph depicts the average percentage of cardiac fusion defects in embryos treated with DMSO or 20 μM LY. The total number of embryos examined from three separate incubations (n) = 45 (DMSO), 45 (20 μM LY). Two-sample t-test; p-value = 4.56 × 10−5. Dots indicate the percent of embryos with cardiac fusion defects per incubation. Letter changes (C, F, I) indicate p-values <0.05. Raw data included in the source file.
-
Figure 1—figure supplement 2—source data 1
Statistical source data for Figure 1—figure supplement 2C, F, I.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp2-data1-v3.xlsx
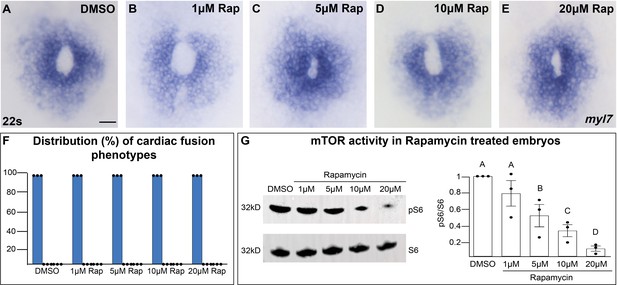
Inhibition of mTOR activity does not affect cardiac fusion.
(A–E) Myocardium visualized with myl7 expression at 22s in embryos treated at bud stage with increasing concentrations of rapamycin (Rap), an inhibitor of mTOR activity. (F) Bar graph displays the distribution of cardiac phenotypes at each rapamycin concentration from three replicates. Total number of embryos analyzed n = 45, 46, 45, 45, 44, respectively. Blue bar = cardiac ring/normal; scale bar = 40 μm. All embryos display cardiac rings, indicating normal cardiac fusion. (G) Representative immunoblot and ratiometric analysis of phosphorylated ribosomal protein S6 (pS6) – a read-out of mTOR activity, to S6 levels reveals that mTOR activity decreases with increasing concentrations of rapamycin. Letter change indicates p < 0.05, one-way analysis of variance (ANOVA). Raw data with full p-values included in the source file.
-
Figure 1—figure supplement 3—source data 1
Statistical source data for Figure 1—figure supplement 3F, G.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp3-data1-v3.xlsx
-
Figure 1—figure supplement 3—source data 2
Original immunoblots used in Figure 1—figure supplement 3G (raw, uncropped) with and without labeling.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp3-data2-v3.zip
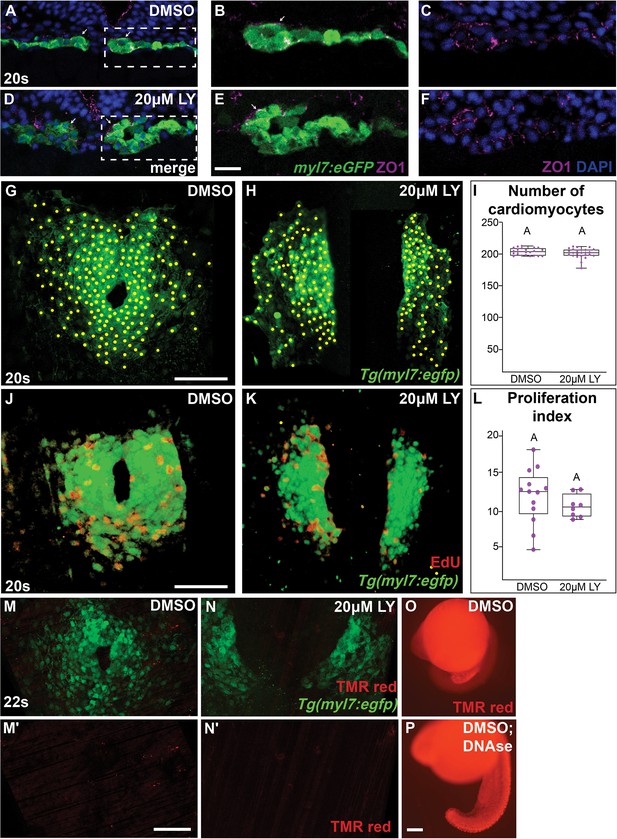
Morphology and proliferation in the myocardium are not compromised in phosphoinositide 3-kinase (PI3K)-inhibited embryos.
Representative transverse cryosections, dorsal to the top, compare the morphology of the myocardium, visualized with Tg(myl7:eGFP) (green), ZO1 (purple), and 4’,6-diamidino-2-phenylindole (DAPI, blue) between DMSO- (A–C) and 20 μM LY- (D–F) treated (bud stage to 20s) embryos. Box (A, D) indicates region magnified in (B, C, E, F). Arrows indicate second dorsal layer. (G–I) Representative images of the myocardium at 20s, which were used to count myocardial cells in DMSO- (G) or 20 μM LY- (H) treated embryos. Yellow dots indicate individual myocardial cells counted using ImageJ. Box-whisker plot displays median number of myocardial cells (I). (J–L) EdU incorporation into the myocardium at 20s in DMSO- (J) and LY- (K) treated embryos following a 1-hr pulse of EdU at 16s. Box-whisker plot displays median proliferation index (L). (M–P) TUNEL staining of Tg(myl7:eGFP) DMSO- and LY-treated embryos (M, N). TUNEL (TMR-red) only channel (M', N'). DMSO only or DMSO and DNAse-treated embryos (O, P). There was no difference in percent of TUNEL+ cardiomyocytes between DMSO- and LY-treated embryos (quantification in source file). n = 21, 25, 13, 8, 17, 19 embryos from 2 to 4 separate bud stage to 20s incubations from (G, H, J, K, M, N), respectively. Scale bars: 10 (A–F), 24 (G, H), 50 (J, K), 60 (M, N), and 10 (O, P) μm. No letter change indicates p > 0.05, two-sample t-test. Raw data and full p-values included in the source file.
-
Figure 1—figure supplement 4—source data 1
Statistical source data for Figure 1—figure supplement 4I, L, M–P.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp4-data1-v3.xlsx

Loss of Pten, an antagonist of phosphoinositide 3-kinase (PI3K) activity, causes cardiac fusion defects.
(A–I) Dorsal views, anterior to the top, of the myocardium labeled with myl7 at 22s. Neither ptena−/− homozygous mutants (A), ptena−/− homozygous; ptenb−/+ heterozygous (B), nor ptena−/−, ptenb−/− double homozygous mutants (C), display cardiac fusion defects. However, maternal contribution of Pten has been reported to persist during development. Adding low concentrations of the Pten inhibitor VO-OHpic (VO-OH – 5, 10 μM) at bud stage to ptena−/−, ptenb−/− double homozygous mutants to inhibit maternal Pten activity did cause a significant increase in cardiac fusion defects (H, I), compared to DMSO (D, G) and low concentrations of VO-OH only (E, F). The number of embryos with cardiac fusion defects and total number analyzed are indicated. (J) Bar graph depicts the distribution of cardiac fusion defects (% of embryos analyzed) in wild-type or ptena−/−, ptenb−/− double homozygous mutants, treated with DMSO, or 5, 10 μM VO-OH. Blue – cardiac ring/normal; orange – fusion only at posterior end. Fisher’s exact test, letter change indicates p < 0.05. Scale bar, 40 μm. Raw data including quantification of all genotypes and full p-values included in the source file.
-
Figure 1—figure supplement 5—source data 1
Statistical source data for Figure 1—figure supplement 5J.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp5-data1-v3.xlsx
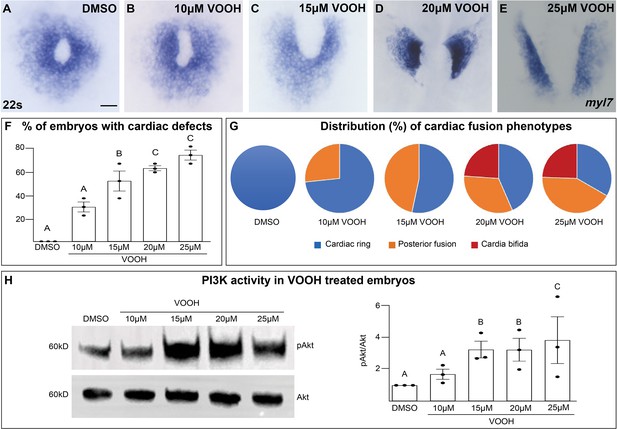
Inhibition of Pten activity with VO-OHpic increases pAkt and causes cardiac fusion defects.
(A–G) Dorsal views of the myocardium labeled with myl7 at 22s in embryos incubated with increasing concentrations of the Pten inhibitor VO-OHpic (VO-OH) from bud stage to 22s. Graphs depicting the average % of embryos displaying cardiac fusion defects (F) and the distribution of cardiac fusion phenotypes (G). Blue – cardiac ring/normal; orange – fusion only at posterior end/mild phenotype, red – cardia bifida/severe phenotype. (H) Representative immunoblot and graph of ratiometric analysis of pAKT to AKT protein levels indicates increasing pAKT levels with increasing concentrations of the Pten inhibitor VO-OH. Three separate incubations per concentration (dots in F, H). n = 15 embryos per incubation per concentration (A–G). Letter change indicates p < 0.05, one-way analysis of variance (ANOVA). Raw data and full p-values included in the source file.
-
Figure 1—figure supplement 6—source data 1
Statistical source data for Figure 1—figure supplement 6F–H.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp6-data1-v3.xlsx
-
Figure 1—figure supplement 6—source data 2
Original immunoblots used in Figure 1—figure supplement 6H (raw, uncropped) with and without labeling.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig1-figsupp6-data2-v3.zip
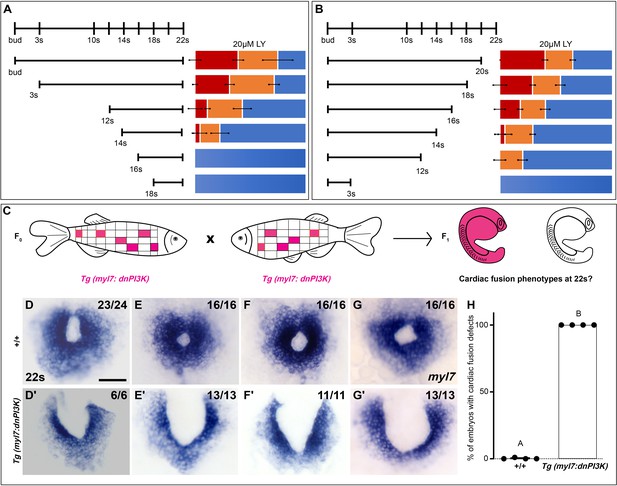
Phosphoinositide 3-kinase (PI3K) is required in the myocardium throughout cardiac fusion.
Graphical representation of the PI3K inhibitor addition (A) and wash-out (B) experiments used to determine the developmental stage over which PI3K is required. In (A) LY is added to embryos at different developmental stages and incubated until 22s, when cardiac fusion is assessed. In (B), LY is added at bud stage and washed-out at different developmental stages, after which embryos are incubated in normal media till 22s, when cardiac fusion is assessed. Bar graphs indicate the average proportion of embryos displaying different phenotypes. Blue – cardiac ring/normal; orange – fusion only at posterior end/mild phenotype, red – cardia bifida/severe phenotype. n = 45 embryos per treatment condition from three biological replicates. (C) Schematic outlines experimental design to test requirement for PI3K in the myocardium. Pink – cells with the Tg(myl7:dnPI3K) transgene. F0 animals are mosaic for the transgene, while all cells in F1 embryos either have the transgene (pink) or do not (white). The myl7 promoter restricts dnPI3K expression to the myocardium in Tg(myl7:dnPI3K) embryos. (D–G) Dorsal view of the myocardium labeled with myl7 in embryos at 22s from four different founder pairs (D–D', E–E', F–F', G–G'). F1 embryos without the Tg(myl7:dnPI3K) transgene (as determined by genotyping) display normal cardiac fusion (D–G, n = 23/24, 16/16, 16/16, 16/16, per founder pair), while F1 siblings with the Tg(myl7:dnPI3K) transgene display cardiac fusion defects (D'–G', n = 6/6, 13/13, 11/11, 13/13), indicating that PI3K signaling is required in myocardial cells. (H) Graph indicating the average % of wild-type and Tg(myl7:dnPI3K)+ embryos with cardiac fusion defects. Letter difference indicates a significant Fisher’s exact test, p = 5.56 × 10−31. Scale bar, 40 μm.
-
Figure 2—source data 1
Statistical source data for Figure 2.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig2-data1-v3.xlsx
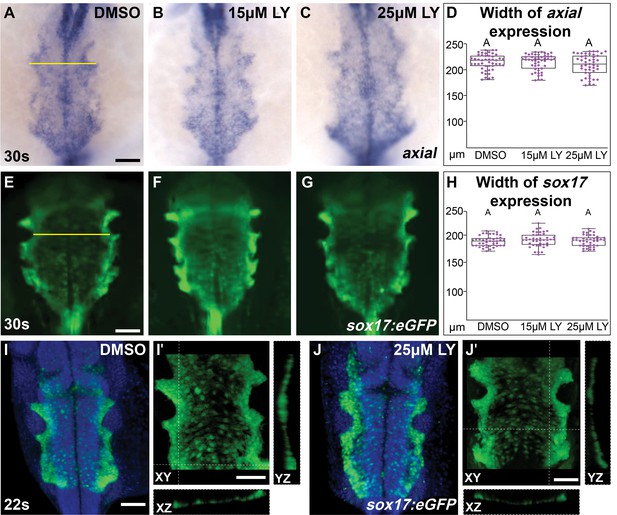
The morphology of endoderm is not compromised in phosphoinositide 3-kinase (PI3K)-inhibited embryos.
Dorsal views, anterior to the top, of the anterior endoderm labeled with axial (A–C) or the Tg(sox17:eGFP) transgene (E–J) at 30s (A–G) or 22s (I, J). Embryos incubated with either DMSO (A, E, I), 15 μM LY (B, F), or 25 μM LY (C, G, J) from the bud stage to 30s (A–H) or 22s (I–J) show no observable difference in the appearance or width of the anterior endoderm. Box-whisker plots display median width of the anterior endoderm from D, H, respectively. n = 47 (axial) and 42 (Tg(sox17:egfp)) embryos per inhibitor concentration from three separate incubations. Yellow lines: width of the endodermal sheet. Purple dots (D, H) indicate individual embryos. No letter differences indicate p-value >0.05 as tested by one-way analysis of variance (ANOVA). High-resolution confocal images of the endoderm at 22s (I, J) further reveals no changes in continuity of the endoderm layer. Three-dimensional reconstructions of the anterior endoderm in DMSO- and 25 μM LY-treated embryos (I, J). Magnifications of I, J with XZ and YZ transverse slices (I’, J’). Tg(sox17:eGFP) transgene = green, blue = DAPI. Scale bars, 60 (A–C), 50 (E–G), 70.6 (I, J) μm. Raw data and full p-values included in the source file.
-
Figure 2—figure supplement 1—source data 1
Statistical source data for Figure 2—figure supplement 1D, H.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig2-figsupp1-data1-v3.xlsx
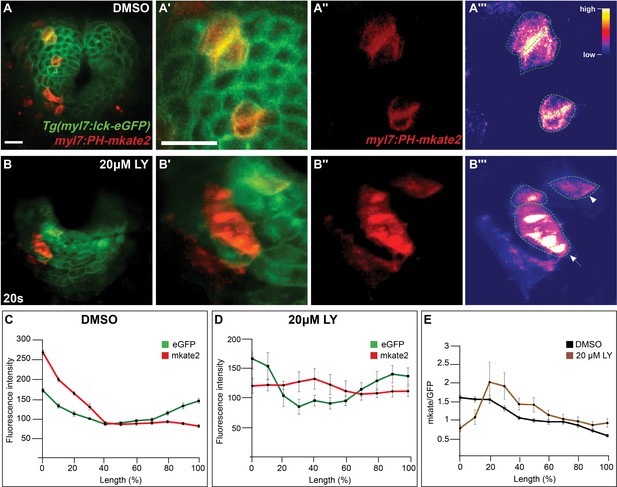
Phosphoinositide 3-kinase (PI3K) activity in myocardial cells.
(A–B) Three-dimensional confocal reconstructions of the myocardium at 20s in DMSO- (A) and LY- (B) treated Tg(myl7:lck-egfp) embryos in which a myl7:PH-mkate2 plasmid was injected to mosaically visualize PI3K activity. PH-mkate2 translocates to the membrane when PI3K activity produces PIP3. In DMSO-treated embryos PH-mkate2 was often found at the membrane enriched asymmetrically (A'–A''') indicating PI3K activity in the myocardium, while in LY-treated embryos PH-mkate2 was defuse in the cytoplasm of myocardial cells (B''' arrowhead) or enriched in subcellular organelles (B''' arrow). A'–A''', B'–B''' are magnifications of A, B, respectively, showing both lck-emgfp and PH-mkate2 (A', B'), PH-mkate2 only (A'', B''), or PH-mkate2 intensities as a heat-map (A''', B'''). (C–E) Graphs depict the average fluorescent intensity for lck-emGFP (green) and PH-mkate2 (red) at different points across the length of a labeled cell, starting with the side of highest mkate2 fluorescence, in DMSO- (C) and LY- (D) treated embryos. The fluorescent intensity of mkate2 normalized to emGFP fluorescence at each point along the line (E) reveals an asymmetrical enrichment of PH-mkate2 at the membrane (0–20%) in DMSO-treated embryos, compared to LY-treated embryos in which PH-mkate2 is enrich in the cytoplasm, the middle of the cell (20–70%). 17 and 13 cells from four DMSO- and four LY-treated embryos were analyzed. Scale bars, 30 μm. Raw data and full p-values included in the source file.
-
Figure 2—figure supplement 2—source data 1
Statistical source data for Figure 2—figure supplement 2C–E.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig2-figsupp2-data1-v3.xlsx
PH-mkate2 is localized asymmetrically at the membrane of myocardial cells in DMSO-treated embryos, but is found in the cytoplasm and subcellular organelles in LY-treated embryos.
Representative time-lapse movie of myocardial cells expressing myl7:PH-mkate2 at 20 s in DMSO- (A) and 20 µM LY- (B) treated embryos. Time-lapse images are a three-dimensional reconstruction of confocal slices taken at 3:30 min intervals, beginning at 20 s.
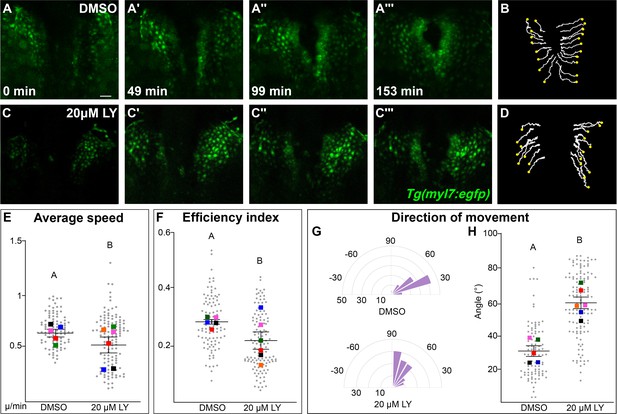
Phosphoinositide 3-kinase (PI3K) signaling regulates the medial movement and speed of the myocardium during cardiac fusion.
Time points from a representative video of myocardial cells visualized with the Tg(myl7:egfp) transgene in embryos treated with DMSO (A, B, Figure 3—video 1) or 20 μM LY (C, D, Figure 3—video 2) from bud stage to 22s. 3D reconstructions of confocal slices (A, C) reveal changes in conformation and location of the myocardium at three major stages of cardiac fusion: early medial movement toward the embryonic midline (A–A', C–C'), posterior merging of bilateral populations (A'', C'') and anterior merging to form a ring (A''', C'''). Representative tracks (B, D) show the paths of a subset of myocardial cells over ~2.5 hr. Yellow dots indicate the starting point of each track. Graphs depict average speed (E), efficiency index (F), and angle of movement (G, H) of myocardial cells. Angular movement along the anterior–posterior axis does not distinguish anterior from posterior movement (G, H). Myocardial cells in PI3K-inhibited embryos show an overall direction of movement that is angular (60–90°) and is slower than in DMSO-treated embryos. 96 and 125 cells were analyzed from five DMSO- and six 20 μM LY-treated embryos, respectively. Gray dots – individual cells; color squares – average per embryo. Average of embryos and standard error (shown in E, F, H). Two-sample t-test, letter change indicates p < 0.05. Scale bars, 60 μm. Quantification details in the methods. Raw data and full p-values included in the source file.
-
Figure 3—source data 1
Statistical source data for quantification of myocardial movement behaviors in Figure 3E–H.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig3-data1-v3.xlsx
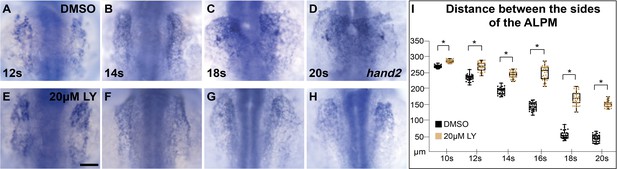
Myocardial movement toward the midline is disrupted in phosphoinositide 3-kinase (PI3K)-inhibited embryos throughout cardiac fusion.
Dorsal views, anterior to the top, of embryos displaying the expression of hand2 in the anterior lateral plate mesoderm (ALPM) at (A, E) 12s, (B, F) 14s, (C, G) 18s, and 20s (D, H), treated with either DMSO (A–D) or 20 μM LY (E–H) at bud stage. (I) Box-whisker plots depict the median distance between the sides of the ALPM. Although, hand2 is properly expressed in LY-exposed embryos, ALPM convergence is affected as early as the 10s stage, with a dramatic difference in convergence starting at 12s. The total number of embryos analyzed (n), from three separate incubations at the noted stages (I) are: n = 34, 33, 31, 32, 34, 34 (DMSO); 32, 29, 30, 34, 31, 28 (20 μM LY), respectively. Dots indicate the distance between ALPM sides per embryo. Student’s t-test: asterisk indicates p-values <0.05. Scale bar, 100 μm. Raw data and full p-values included in the source file.
-
Figure 3—figure supplement 1—source data 1
Statistical source data for distance between bilateral anterior lateral plate mesoderm (ALPM) populations, Figure 3—figure supplement 1I.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig3-figsupp1-data1-v3.xlsx

Phosphoinositide 3-kinase (PI3K) signaling directs myocardial movement during the early stages of cardiac fusion and regulates velocity along the medial-lateral axis.
(A, B) Time-lapse confocal reconstructions from Figure 3 overlaid with cell movement tracks, starting at t = 0 (yellow dots). Graphs of average speed (C) and direction of movement (D) subdivided into three distinct stages of cardiac fusion: early movement (0–48min), posterior fusion (49–99min), and anterior fusion (100–153min). Graphs of average absolute medial–lateral velocity (E) and average absolute angular velocity (F) sub-divided into cells at the top, middle, and bottom of the myocardial populations. The average speed of myocardial cells in LY-treated embryos is consistently slower than DMSO-treated embryos (C). This defect appears to derive mostly from differences in medial-lateral movement (E) rather than in angular movement (F). In LY-treated embryos, myocardial cells display a more angular average direction of movement compared to DMSO-treated embryos during the early stage of cardiac fusion (early movement), after which wild-type myocardial cell movement becomes more angular matching myocardial cell movements in LY-treated embryos (D). Gray dots – individual cells, color squares – average per embryo. Average and standard error based on embryos. Scale bar, 60μm. Letter change indicates p < 0.05, one-way analysis of variance (ANOVA) (C, D, F) or t-test (E). Raw data and full p-values included in the source file.
-
Figure 3—figure supplement 2—source data 1
Statistical source data for quantification of myocardial movement separated by stages of movement and location, Figure 3—figure supplement 2C–F.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig3-figsupp2-data1-v3.xlsx
Myocardial cells in DMSO-treated embryos collectively move toward the midline and form a ring during cardiac fusion.
Representative time-lapse movie of cardiac fusion in a DMSO-treated embryo. Myocardial cells visualized with Tg(myl7:egfp) (A), tracks show movement of selected cells (B), and overlay of eGFP and tracks (C). Time-lapse images are of three-dimensional reconstruction of confocal slices taken at 4:32 min intervals for 2.5 hr, beginning at 14s.
Myocardial cells in phosphoinositide 3-kinase (PI3K)-inhibited embryos fail to move properly toward the midline.
Representative time-lapse movie of myocardial cells visualized with Tg(myl7:egfp) (A), tracks of selected cells (B), and overlay of tracks and eGFP (C) from an embryo treated with 20 μM LY from bud-20s. Time-lapse was acquired as described in Figure 3—video 1.
Phosphoinositide 3-kinase (PI3K) signaling promotes the medial directional movement of myocardial cells toward the midline.
Side-by-side comparison of myocardial movement in DMSO- (A, Figure 3—video 1) and LY- (B, Figure 3—video 2) treated embryos reveals that inhibition of PI3K signaling by LY prevents myocardial cells from being adequately directed toward the midline. Selected analyzed tracks (white lines) overlaying 3D reconstructions of the Tg(myl7:egfp) transgene (green) in DMSO- (A) and 20 μM LY- (B) treated embryos.
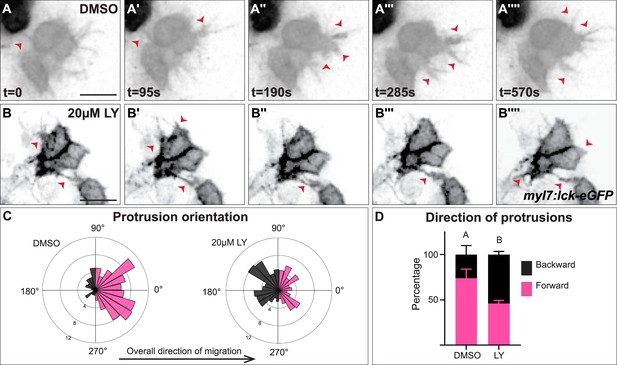
Myocardial membrane protrusions are misdirected in phosphoinositide 3-kinase (PI3K)-inhibited embryos.
(A–B'''') timepoints from representative videos (see Figure 4—video 1) of myocardial cells whose membrane has been labeled with myl7:lck-eGFP (black), medial to the right, in a DMSO- (A–A'''') or a 20 μM LY- (B–B'''') treated embryo. Red arrowheads indicate representative protrusions, which are mostly oriented medially, coincident with the direction of movement in DMSO-treated embryos (A–A'''') but are oriented in all directions in LY-treated embryos (B–B''''). Rose (C) and bar (D) graphs displaying the orientation of membrane protrusions in DMSO- (left) or LY- (right) treated embryos. The length of each radial bar in (C) represents the percentage of protrusions in each bin. Bar graph displays the total percentage of forward or backward protrusions. Forward protrusions: 270–90°, pink. Backward protrusions: 90–270°, black. n = 425 protrusions from 11 cells in 5 embryos (DMSO), and 480 protrusions from 11 cells in 4 embryos (20 μM LY). Fisher’s exact test, p-value 1.8 × 10−5. Mean ± standard error. Scale bar, 30 μm. Raw data included in the source file.
-
Figure 4—source data 1
Statistical source data for quantification of myocardial protrusion properties.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig4-data1-v3.xlsx
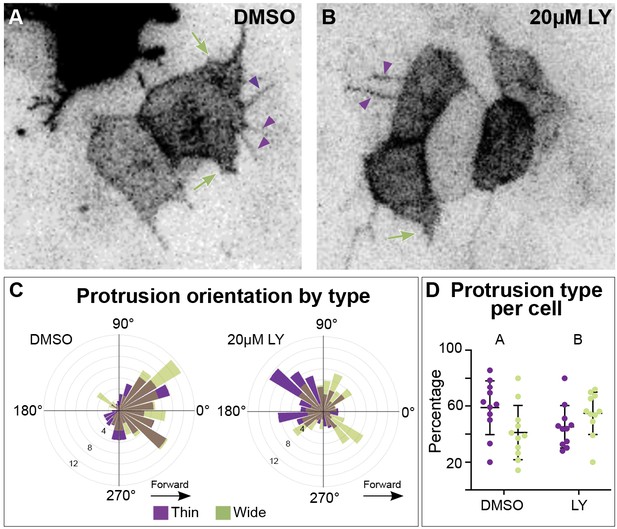
Different types of myocardial protrusion morphologies occur during cardiac fusion.
Snapshots from timelapse videos of myocardial cells mosaically labeled with myl7:lck-eGFP in a DMSO- (A) or a LY- (B) treated embryo. Purple arrowheads and green arrows indicate thin and wide protrusions, respectively. (C) Rose plots display the orientation of thin and wide protrusions in DMSO- (left) or LY- (right) treated embryos. Forward/medial direction: 270–90°. Backward/lateral direction: 90–270°. (D) Graphs indicate the average percentage of thin (purple dots) or wide (green dots) protrusions per cell per hour. Dots = individual cells. n = 425 protrusions from 11 cells in 5 embryos (DMSO), and 480 protrusions from 11 cells in 4 embryos (20 µM LY). Fisher’s exact test, p-value 3.866e-08. Mean ± standard deviation. Raw data included in the source file.
-
Figure 4—figure supplement 1—source data 1
Statistical and raw source data for Figure 4—figure supplement 1C, D.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig4-figsupp1-data1-v3.xlsx
Dynamic medially oriented myocardial membrane protrusions are lacking in phosphoinositide 3-kinase (PI3K)-inhibited embryos.
Representative time-lapse movies of myocardial membrane protrusions during cardiac fusion, visualized by injecting myl7:lck-eGFP plasmids at the one-cell stage, in DMSO- (left panel) or 20 μM LY- (right panel) treated embryos. Left panel highlights membrane protrusions (red arrowheads) in a set of posterior myocardial cells in a DMSO-treated embryo. Myocardial membrane protrusions in DMSO-treated embryos are mostly directed in the medial orientation (toward the right in both panels). Right panel highlights myocardial membrane protrusions (red arrowheads) in PI3K-inhibited embryos during cardiac fusion. Medial membrane protrusions (toward the right) are lacking in PI3K-inhibited embryos. DMSO or LY treatment from bud to 20s. Time-lapse movies are 3D reconstruction of confocal images of membrane protrusions taken at ~90-s intervals for 2 hr. Scale bar, 10 μm.
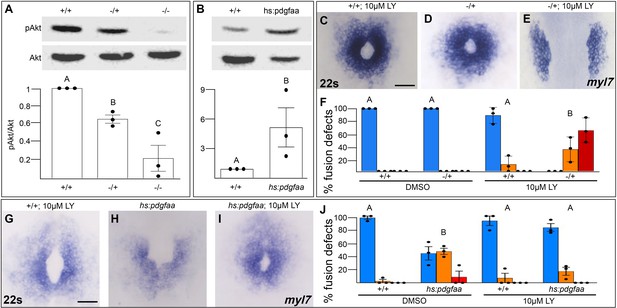
Pdgfra activates and genetically interacts with phosphoinositide 3-kinase (PI3K) signaling to regulate cardiac fusion.
(A, B) Immunoblot and ratiometric analysis of phosphorylated Akt (pAkt) compared to total Akt levels reveals reduced pAkt levels in loss-of-function pdgfrask16 heterozygous (−/+) or homozygous (−/−) mutant embryos at 22s (A), and elevated pAkt levels at 22s when PDGF signaling is activated with the Tg(hs:pdgfaa) transgene (B). Bar graphs display averages from three separate experiments. (C–E, G–I) Dorsal views, anterior to the top, of the myocardium labeled with myl7 at 22s. In contrast to a normal ring of myocardial cells found in wild-type embryos treated with 10 μM LY starting at bud stage (C) or in pdgfra heterozygous embryos (D), when pdgfra heterozygous mutants are exposed to 10 μM LY, cardiac fusion is defective with embryos displaying severe phenotypes such as cardia bifida (E). Furthermore, the percent of cardiac fusion defects observed in Tg(hs:pdgfaa) embryos heat-shocked at bud stage (H) is greatly decreased when heat-shocked Tg(hs:pdgfaa) embryos are exposed to 10 μM LY at bud stage (I). (F, J) Bar graphs depict the average distribution of cardiac fusion defects from the indicated genotypes. The total number of embryos examined over three separate replicates are 47 (DMSO, +/+), 25 (DMSO, −/+), 36 (10 μM LY, +/+), 31 (10 μM LY, −/+), 49 (heat-shock, DMSO, +/+), 57 (heat-shock, DMSO, Tg(hs:pdgfaa)), 51 (heat-shock, 10 μM LY, +/+), 57 (heat-shock, 10 μM LY, Tg(hs:pdgfaa)). Blue – cardiac ring/normal; orange – fusion only at posterior end/mild, red – cardia bifida/severe. Bar graphs, mean ± standard error. One-way analysis of variance (ANOVA) (A, F, J) or Student’s t-test (B), letter change indicates p < 0.05. Scale bar, 60 μm. Raw data and full p-values included in the source file.
-
Figure 5—source data 1
Statistical source data for Figure 5A, B, F, J.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Original immunoblots used in Figure 5A, B (raw, uncropped) with and without labeling.
- https://cdn.elifesciences.org/articles/85930/elife-85930-fig5-data2-v3.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Danio rerio) | Tg(myl7: eGFP)twu34 | Huang et al., 2003 | twu34; RRID: ZFIN_ZDB-GENO-050809-10 | Transgenic |
| Genetic reagent (Danio rerio) | Tg(sox17:eGFP)ha01 | Mizoguchi et al., 2008 | ha01; RRID: ZFIN_ZDB-GENO-080714-2 | Transgenic |
| Genetic reagent (Danio rerio) | Tg(hsp70l:pdgfaa-2A-mCherry;cryaa:CFP)sd44 | Bloomekatz et al., 2017 | sd44; ZDB-ALT-170425-5 | Transgenic |
| Genetic reagent (Danio rerio) | ref (pdgfrask16) | Bloomekatz et al., 2017 | sk16; ZDB-ALT-170329-1 | Mutant |
| Genetic reagent (Danio rerio) | ptenahu1864 | Faucherre et al., 2008 | hu1864; ZDB-ALT-080910-1 | Mutant |
| Genetic reagent (Danio rerio) | ptenbhu1435 | Faucherre et al., 2008 | hu1435; ZDB-ALT-080910-2 | Mutant |
| Genetic reagent (Danio rerio) | Tg(myl7:dnPI3K; Cryaa:CFP) | This paper | Transgenic, see Materials and methods | |
| Genetic reagent (Danio rerio) | Tg(myl7:lck-emgfp) | This paper | Transgenic, see Materials and methods | |
| Recombinant DNA reagent | pBSRN3-∆p85 | Carballada et al., 2001 | ||
| Other | myl7 | Yelon et al., 1999 | ZDB-GENE-991019-3 | mRNA probe |
| Other | axial | Strähle et al., 1993 | ZDB-GENE-980526-404 | mRNA probe |
| Other | hand2 | Yelon et al., 2000 | ZDB-GENE-000511-1 | mRNA probe |
| Chemical compound, drug | LY294002 | Millipore-Sigma | Cat# 154447-36-6 | |
| Chemical compound, drug | Dactolisib | Millipore-Sigma | Cat# 915019-65-7 | |
| Chemical compound, drug | Pictilisib | Millipore-Sigma | Cat# 957054-30-7 | |
| Chemical compound, drug | Rapamycin | Selleckchem | Cat# S1039 | |
| Chemical compound, drug | VO-Ohpic trihydrate | Selleckchem | Cat# S8174 | |
| Antibody | phospho-AKT (Rabbit monoclonal) | Cell Signaling | Cat# 4060, RRID: AB_2315049 | WB(1:2000) |
| Antibody | pan-AKT (Rabbit monoclonal) | Cell Signaling | Cat# 4691, RRID: AB_915783 | WB (1:2000) |
| Antibody | Anti-rabbit IgG HRP-conjugated (Goat polyclonal) | Cell Signaling | Cat# 7074, RRID: AB_2099233 | WB (1:5000) |
| Antibody | anti-GFP (Chicken polyclonal) | Abcam | Cat# ab13970, RRID: AB_300798 | IF (1:1000) |
| Antibody | anti-ZO-1 (Mouse monoclonal) | Thermo Fisher Scientific | Cat# 33–9100, RRID: AB_87181 | IF (1:200) |
| Antibody | anti-chicken-488 (Goat polyclonal) | Thermo Fisher Scientific | Cat# A32931TR, RRID: AB_2866499 | IF (1:300) |
| Antibody | anti-mouse-647 (Goat polyclonal) | Thermo Fisher Scientific | Cat# A32728, RRID: AB_2633277 | IF (1:300) |
| Commercial assay or kit | Cell Death detection kit, TMR red | Millipore Sigma | Cat# 12156792910 | |
| Commercial assay or kit | Click-&-Go Cell Proliferation Assay Kit | Click Chemistry Tools | Cat# 1328 | |
| Software, algorithm | mTrackJ | Meijering et al., 2012 | ImageJ Plugin for motion tracking and analysis | |
| Software, algorithm | Correct 3D Drift | Parslow et al., 2014 | ImageJ Plugin for sample drift correction | |
| Software, algorithm | Prism 10.0.2 | GraphPad | RRID: SCR_002798 | Data visualization and statistics software |
| Software, algorithm | Leica | LASX | Leica Application Suite X, RRID: SCR_013673 | Microscope image processing software |
Primers for genotyping and cloning.
| Name | Sequence (5′–3′) | |
|---|---|---|
| Primers to screen for the Tg(myl7:dnPI3K) transgene in F1 embryos | dnPI3K_F1 | gcgggaagaggacattgact |
| dnPI3K_R1 | gcgggaagaggacattgact | |
| Primers to clone lck-emGFP into the middle-entry vector of the tol2 gateway system | Hi_lck_1F | cagtcgactggatccggtacagatccgctagccaccatg |
| Hi_lck_1R | cagtcgactggatccggtacagatccgctagccaccatg | |
| Hi_emgfp_2F | ggtcgccaccgtgtccaagggcgaggag | |
| Hi_emgfp_2R | ggtcgccaccgtgtccaagggcgaggag | |
| Primers to replace actc1b promoter with myl7 promoter in Addgene plasmid 109501 | Hi_CbPHmkate2_F | ggctgaaaagcaatcctgcagtgaccaaagcttaaatcagttg |
| Hi_CbPHmkate2_R | ctctccagaatcactgcggccatggccatggtggctacggatc |
| Inhibitors | Target | Known off-targets |
|---|---|---|
| LY294002 | Class I PI3Ks | mTOR, DNA-PK |
| Dactolisib | Class I PI3Ks, mTOR | |
| Pictilisib | Class I PI3Ks | mTOR |
| Rapamycin | mTOR | |
| VOOH | Pten |





