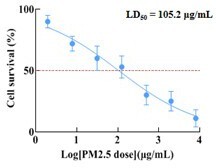
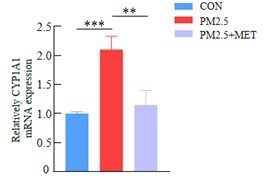
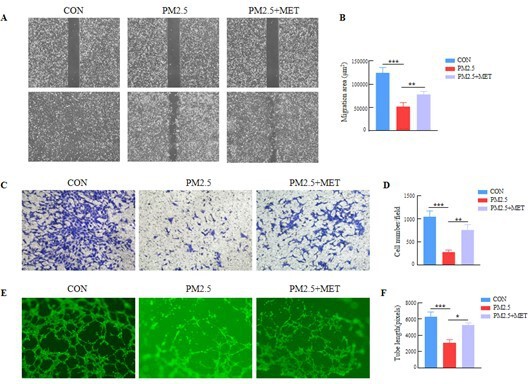
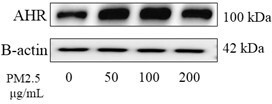
PM2.5 leads to adverse pregnancy outcomes by inducing trophoblast oxidative stress and mitochondrial apoptosis via KLF9/CYP1A1 transcriptional axis
Figures

The geographic location of the PM2.5 collection site and analysis of PM2.5 morphological and elemental composition.
(A) Map and coordinates of the PM2.5 sampling site. The image was obtained from Google Maps. (B) SEM images of PM2.5 in an enlarged field. Scale bar, 50 μm (left image), 10 μm (middle image) and 5 μm (right image). (C) Analysis of PM2.5 elemental composition via SEM coupled with EDS.
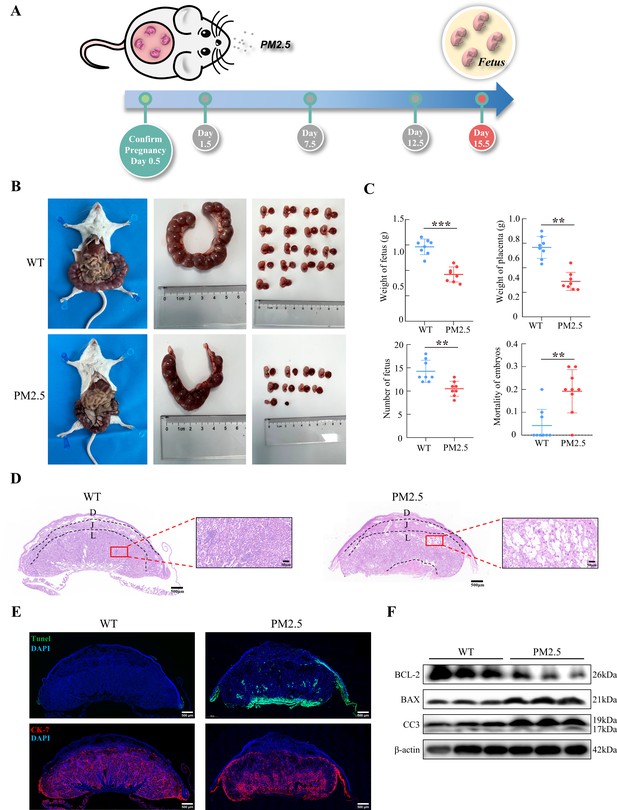
Gestational PM2.5 exposure resulted in adverse pregnancy outcomes and impaired placental development in mice.
(A) Schematic diagram of the in vivo exposure model constructed by intratracheal instillation of PM2.5 in mice at 1.5 d, 7.5 d, and 12.5 d of pregnancy. (B) Representative images of uterine, fetal, and placental morphology in wild-type and PM2.5 exposed mice at 15.5 d of gestation. (C) The effect of PM2.5 on the fetus and placenta, including the weight of the fetus, the weight of the placenta, the number of the fetuses, and the mortality of the embryos (n=8, the dots in the top two graphs indicate the average placental or fetal mouse weight per pregnant mouse) (**, p<0.01; ***, p<0.001). (D) Representative HE staining of placental tissues from the wild-type and PM2.5-exposed mice. Compared with the wild-type placental tissues, the labyrinth layer in the PM2.5-exposed group was loosely distributed and disorganized, with a large number of vacuolated structures and large intercellular gaps (D: Decidual; J: Junctional zone; L: Labyrinth). Scale bar, 500 μm (upper images) and 50 μm (lower images) (E) Apoptosis of placental cells was detected by TUNEL staining. TUNEL: apoptotic cells; DAPI: nuclei; CK-7: trophoblast cells. Scale bar, 500 μm. (F) The expression of apoptosis-related proteins in the placental tissue of mice was detected by western blotting. PM2.5 decreased the expression of the anti-apoptotic protein BCL-2 and increased the expression of the pro-apoptotic proteins BAX and Cleaved-caspase 3 (CC3).
-
Figure 2—source data 1
Labelled gel images.
(Figure 2F-BCL-2) The expression of BCL-2 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-BAX) The expression of BAX expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-CC3) The expression of Cleaved-Caspase 3 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig2-data1-v2.zip
-
Figure 2—source data 2
Raw unlabelled gel images.
(Figure 2F-BCL-2) The expression of BCL-2 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-BAX) The expression of BAX expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-CC3) The expression of Cleaved-Caspase 3 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 2F-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig2-data2-v2.zip

PM2.5 impaired the biological functions of HTR8/SVneo.
(A) Representative TEM images of morphological changes of HTR8/SVneo caused by PM2.5. The green arrows indicated mitochondria in the cell. Scale bar, 2 μm (left images) and 1 μm (right images) (B) Detection of PM2.5 particles internalized by HTR8/SVneo cells using flow cytometry. The horizontal coordinates of the FSC-A represented the size of the particles, and the vertical coordinates of the SSC-A represented intracellular particle complexity. (FSC-A: Forward Scatter-Area; SSC-A: Side Scatter-Area) (C) Proliferation curves were determined by CCK-8 assays. The plots represented the proliferative ability of HTR8/SVneo cells. (D) Representative images showing the suppressed proliferation of HTR8/SVneo cells by PM2.5. The nuclei were stained with DAPI (blue), the cells in the proliferation stage were stained with EDU (red). Scale bar, 100 μm. (E) Quantitative analysis of the proliferation rate of HTR8/SVneo cells. (F) Annexin V-FITC/PI staining and flow cytometry analysis were used to determine the percentage of apoptotic cells following treatment with different concentrations of PM2.5. (G) Histogram analysis indicated the apoptotic rate of the cells in each group. (H) Representative images of the wound healing assay of HTR8/SVneo cells at 0 and 24 hr time points following treatment with different concentrations of PM2.5. Scale bar, 200 μm. (I) The histogram indicated the migration area (μm2) in each group. (J) The representative images of Transwell invasion assay at 24 h in the different groups. Scale bar, 50μm. (K) The histogram indicated the proportion of cells in each group. (L) Representative images showing cell tube formation following treatment with different concentrations of PM2.5 for 4 hr. Scale bar, 100μm. (M) The histogram indicated the comparison of the tube length of HTR8/SVneo cells (*, p<0.05; **, p<0.01; ***, p<0.001).
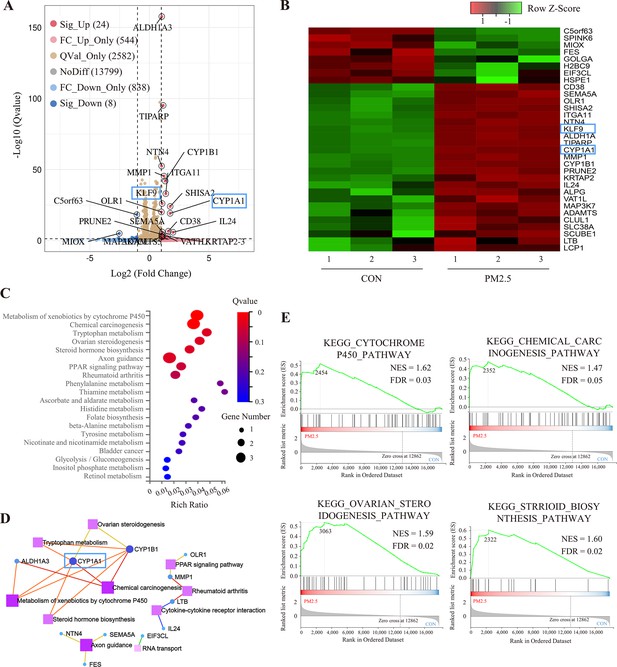
RNA sequencing and bioinformatic analysis of PM2.5-induced genomic alterations in HTR8/SVneo cells.
(A) Volcano plot of the differentially expressed genes (DEGs) in the RNA-Seq (Qvalue <0.05; |Log2 (fold change) |>1). (B) Heat map visualization of sequencing data with the Row Z-Score scaling method for gene expression between samples. (C) Bulb map of KEGG analysis for the DEGs. The rich ratio represented the enrichment degree of DEGs. (D) Topological networks based on altered gene enrichment analysis. (E) GSEA of RNA-Seq data showed the enrichment of genes in the pathway. The zero-cross line indicates the point in which the difference between expression in the PM2.5-treated and control groups is zero. NES, normalized enrichment score; FDR, false discovery rate.
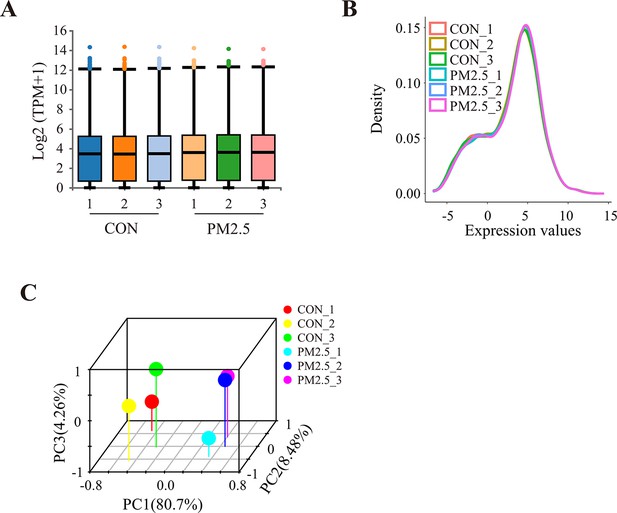
RNA sequencing and bioinformatic analysis.
(A) The boxplots showed the distribution of gene expression levels in the control and exposed groups. (B) Density plots for the distribution of gene expression in different sample groups. (C) 3D principal component analysis scatter plots of HTR8/SVneo cells following the different interventions.

PM2.5 impaired the biological functions of HTR8/SVneo cells by triggering oxidative stress.
(A) Detection of GSH, SOD, and MDA expression levels in the placenta of wild-type and PM2.5 exposed mice (n=8, one randomly selected placental sample per mouse). (B) Expression of antioxidant-related proteins HO-1, NQO-1, GCLC, and SOD-1 in the placenta of wild-type and PM2.5-exposed mice by western blotting (n=3). (C) Expression of antioxidant-related proteins HO-1, NQO-1, GCLC, and SOD-1 in HTR8/SVneo cells treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, and 200 μg/mL) by western blotting. (D) Detection of GSH, SOD, and MDA expression levels in HTR8/SVneo treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). (E) Detection of intracellular ROS levels by DCFH-DA probe staining following treatment with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). The nuclei were counter stained with Hoechst (blue), and ROS were stained with DCFH-DA (green). Scale bar, 50 μm. (F) Quantitative detection of intracellular ROS levels by flow cytometry after staining with DCFH-DA. The histogram indicated the mean levels of FITC fluorescence in each group. (G) Mitochondrial ROS levels in HTR8/SVneo treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). Hoechst was used to counterstain the nuclei; Mito-tracker was used to stain the mitochondria; Mito-SOX was used to stain the mitochondrial ROS. Scale bar, 50 μm. (H) The histogram indicated the Mito-SOX Red Staining mean intensity in each group. (I) Mitochondrial membrane potential was assessed using JC-1 staining following treatment with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). When the mitochondrial membrane potential was high, JC-1 aggregated in the mitochondrial matrix, forming J-aggregates, which fluoresced red; when the mitochondrial membrane potential was low, JC-1 did not aggregate in the mitochondrial matrix, thus, JC-1 was present as a monomer, which fluoresced green. Scale bar, 50 μm. (J) The histogram indicated the Red/Green intensity in each group. (K) Expression of mitochondrial apoptosis-associated proteins in the mitochondria and cytoplasm using western blotting following treatment with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). Cytochrome C (Cyt-C) expression was detected in the mitochondria, while Cyt-C, BCL-2, BAX and Cleaved-caspase 3 (CC3) expression levels in the cytoplasm were detected. The mitochondrial marker VDAC and the cytosol marker β-actin were used to identify the purity of the mitochondria in the extracts (*, p<0.05; **, p<0.01; ***, p<0.001).
-
Figure 5—source data 1
Labelled gel images.
(Figure 5B-HO-1) The expression of HO-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns) (Figure 5B-NQO-1). The expression of NQO-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns) (Figure 5B-GCLC). The expression of GCLC expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns) (Figure 5B-SOD-1). The expression of SOD-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns) (Figure 5B-β-actin). The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns) (Figure 5C-HO-1). The expression of HO-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5C-NQO-1). The expression of NQO-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5C-GCLC). The expression of GCLC expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5C-SOD-1). The expression of SOD-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5C-β-actin). The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Mito-Cyt-c). The expression of Cyt-c expression in mitochondria after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Mito-VDAC-1). The expression of VDAC-1 expression in mitochondria after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Cyto-Cyt-c). The expression of Cyt-c expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Cyto-BCL-2). The expression of BCL-2 expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Cyto-BAX). The expression of BAX expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Cyto-CC3). The expression of Cleaved-Caspase 3 expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL) (Figure 5K-Cyto-β-actin). The expression of β-actin expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig5-data1-v2.zip
-
Figure 5—source data 2
Raw unlabelled gel images.
(Figure 5B-HO-1) The expression of HO-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 5B-NQO-1) The expression of NQO-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 5B-GCLC) The expression of GCLC expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 5B-SOD-1) The expression of SOD-1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 5B-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 5C-HO-1) The expression of HO-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5C-NQO-1) The expression of NQO-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5C-GCLC) The expression of GCLC expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5C-SOD-1) The expression of SOD-1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5C-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Mito-Cyt-c) The expression of Cyt-c expression in mitochondria after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Cyto-Cyt-c) The expression of Cyt-c expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Cyto-BAX) The expression of BAX expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Cyto-CC3) The expression of Cleaved-Caspase 3 expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 5K-Cyto-β-actin) The expression of β-actin expression in cytoplasm after treated with different concentration of PM2.5 (0, 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig5-data2-v2.zip

NAC reversed PM2.5-induced impairment of HTR8/SVneo cell function by inhibiting oxidative stress.
(A) GSH, SOD, and MDA expression levels in HTR8/SVneo cells treated with PM2.5 (100μg/mL) and NAC (5 mM). (B) Intracellular ROS levels following treatment with PM2.5 (100 μg/mL) and NAC (5 mM). The nuclei were stained blue with Hoechst, the intracellular ROS in cells were stained green with DCFH-DA. Scale bar, 50 μm. (C) Quantitative detection of intracellular ROS levels by flow cytometry after staining with DCFH-DA. The histogram indicated the mean FITC fluorescence of each group (100 μg/mL PM2.5 and 5 mM NAC). (D) Images showing the proliferation of the HTR8/SVneo cells treated with PM2.5 (100 μg/mL) and NAC (5 mM) by EDU assay. The nuclei were stained with DAPI (blue), and the proliferating cells were stained with EDU (red). Scale bar, 100 μm. (E) Quantitative analysis of the proliferation rate of HTR8/SVneo cells. (F) Detection of the percentage of apoptotic cells treated with PM2.5 (100 μg/mL) and NAC (5 mM) using flow cytometry assay. (G) The histogram analysis showed the apoptotic rate of cells in each group. (H) Representative images of the wound healing assay of HTR8/SVneo cells at the 0 and 24 hr. Scale bar, 200 μm. (I) The histogram showed the migration area (μm2) in each group. (J) Representative images of the Transwell invasion assays after 24 hr in the different groups. Scale bar, 50 μm (K) The histogram showed the cell counts/field in each group. (L) Representative images showing tube formation at different concentrations of PM2.5 after 4 hr. Scale bar, 100 μm. (M) The histogram showed the quantification of tube length of HTR8/SVneo cells. (N) The protein expression of BCL-2, BAX, Cleaved-Caspase3 by western blotting (*, p<0.05; **, p<0.01; ***, p<0.001).
-
Figure 6—source data 1
Labelled gel images.
(Figure 6N-BCL-2) The expression of BCL-2 expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-BAX) The expression of BAX expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-CC3) The expression of Cleaved-Caspase 3 expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-β-actin) The expression of β-actin expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig6-data1-v2.zip
-
Figure 6—source data 2
Raw unlabelled gel images.
(Figure 6N-BCL-2) The expression of BCL-2 expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-BAX) The expression of BAX expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-CC3) The expression of Cleaved-Caspase 3 expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC). (Figure 6N-β-actin) The expression of β-actin expression in HTR8/SVneo (CON, PM2.5, PM2.5+NAC).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig6-data2-v2.zip
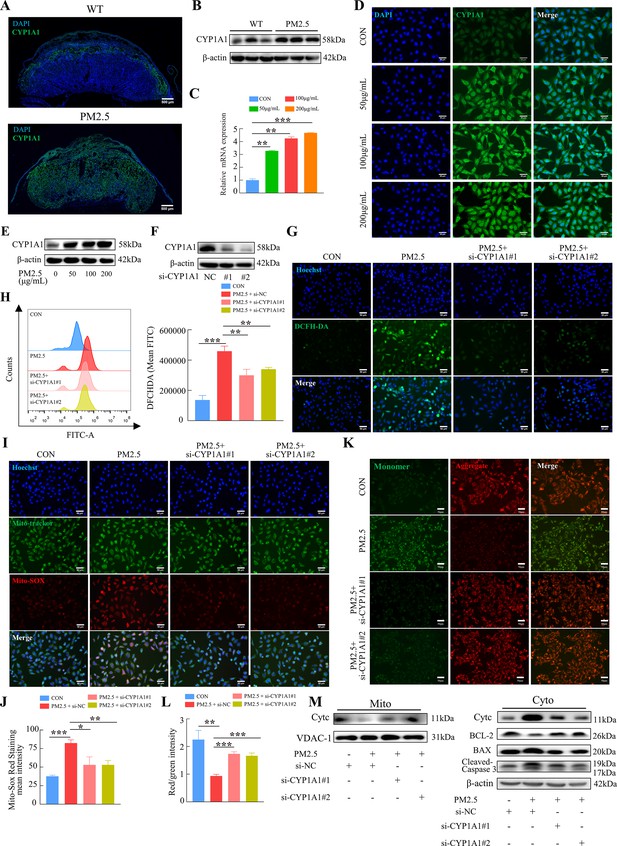
PM2.5 caused mitochondrial apoptosis through oxidative stress damage in HTR8/SVneo cells via CYP1A1.
(A) Immunofluorescence staining of CYP1A1 in mice placental tissue sections. The nuclei were stained with DAPI (blue). The CYP1A1 expressing cells were stained with a specific antibody (green). Scale bar, 500 μm. (B) The protein expression levels of CYP1A1 in the placental tissues were detected by western blotting (n=3). (C) Relative mRNA expression levels of CYP1A1 in HTR8/SVneo cells were treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). (D) Immunofluorescence images of CYP1A1 in HTR8/SVneo cells were treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). The nuclei were stained with DAPI (blue), and CYP1A1 was stained with specific antibody (green). Scale bar, 50 μm. (E) The protein expression levels of CYP1A1 in HTR8/SVneo cells treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). (F) The effects of CYP1A1 knockdown using two siRNAs by western blotting. (G) Quantitative detection of intracellular ROS by flow cytometry after staining with DCFH-DA. The histogram showed the mean FITC in each group. (H) Representative images of DCFH-DA staining for intracellular ROS (PM2.5: 100 μg/mL). The nuclei were stained with Hoechst (blue), and the ROS in cells were stained with DCFH-DA (green). Scale bar, 50 μm. (I) Mitochondrial ROS levels in HTR8/SVneo (PM2.5: 100 μg/mL). Hoechst was used to label the nuclei of cells in blue; Mito-tracker was used to label the mitochondrial sites; Mito-sox was used to label the mitochondrial ROS. Scale bar, 50 μm. (J) The histogram indicated the Mito-SOX Red Staining mean intensity in each group. (K) Detection of mitochondrial membrane potential by JC-1 staining (PM2.5: 100 μg/mL). Scale bar, 50 μm. (L) The histogram indicated the Red/Green intensity in each group. (M) Mitochondrial apoptosis-associated protein expression in mitochondria and cytoplasm using western blotting (PM2.5: 100 μg/mL). Cyt-C expression was detected in mitochondria, and Cyt-C, BCL-2, BAX and Cleaved-caspase 3 (CC3) expression levels in the cytoplasm were detected. The mitochondrial marker VDAC and the cytosol marker β-actin were used to identify the purity of the mitochondria in the extracts. (si-CYP1A1#1, si-CYP1A1#2: siRNAs to knockdown CYP1A1; si-NC: siRNAs Negative Control; *, p<0.05; **, p<0.01; ***, p<0.001).
-
Figure 7—source data 1
Labelled gel images.
(Figure 7B-CYP1A1) The expression of CYP1A1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 7B-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 7E-CYP1A1) The expression of CYP1A1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7E-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7F-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo cells (si-NC, si-CYP1A1#1, si-CYP1A1#2). (Figure 7F-β-actin) The expression of β-actin expression in the HTR8/SVneo cells (si-NC, si-CYP1A1#1, si-CYP1A1#2). (Figure 7M-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig7-data1-v2.zip
-
Figure 7—source data 2
Raw unlabelled gel images.
(Figure 7B-CYP1A1) The expression of CYP1A1 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 7B-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 7E-CYP1A1) The expression of CYP1A1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7E-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7F-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo cells (si-NC, si-CYP1A1#1, si-CYP1A1#2). (Figure 7F-β-actin) The expression of β-actin expression in the HTR8/SVneo cells (si-NC, si-CYP1A1#1, si-CYP1A1#2). (Figure 7M-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2). (Figure 7M-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- CYP1A1#1, PM2.5+si- CYP1A1#2).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig7-data2-v2.zip
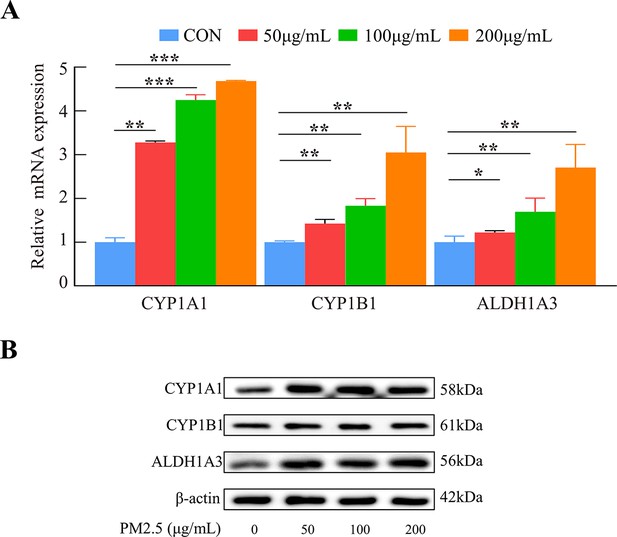
Gene expression in HTR8/SVneo cells under PM2.5 treatment.
(A) CYP1A1, CYP1B1, and ALDH1A3 mRNA with different concentrations of PM2. 5 (50 μg/mL, 100 μg/mL, 200 μg/mL) were measured by qRT-PCR. (B) The protein expression of CYP1A1, CYP1B1, and ALDH1A3 in HTR8/SVneo treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). (*, p<0.05; **, p<0.01; ***, p<0.001).
-
Figure 7—figure supplement 1—source data 1
Labelled gel images.
(Figure 7—figure supplement 1B–CYP1A1) The expression of CYP1A1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B–CYP1B1) The expression of CYP1B1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B–ALDH1A3) The expression of ALDH1A3 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig7-figsupp1-data1-v2.zip
-
Figure 7—figure supplement 1—source data 2
Raw unlabelled gel images.
(Figure 7—figure supplement 1B–CYP1A1) The expression of CYP1A1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B–CYP1B1) The expression of CYP1B1 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B–ALDH1A3) The expression of ALDH1A3 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 7—figure supplement 1B-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig7-figsupp1-data2-v2.zip
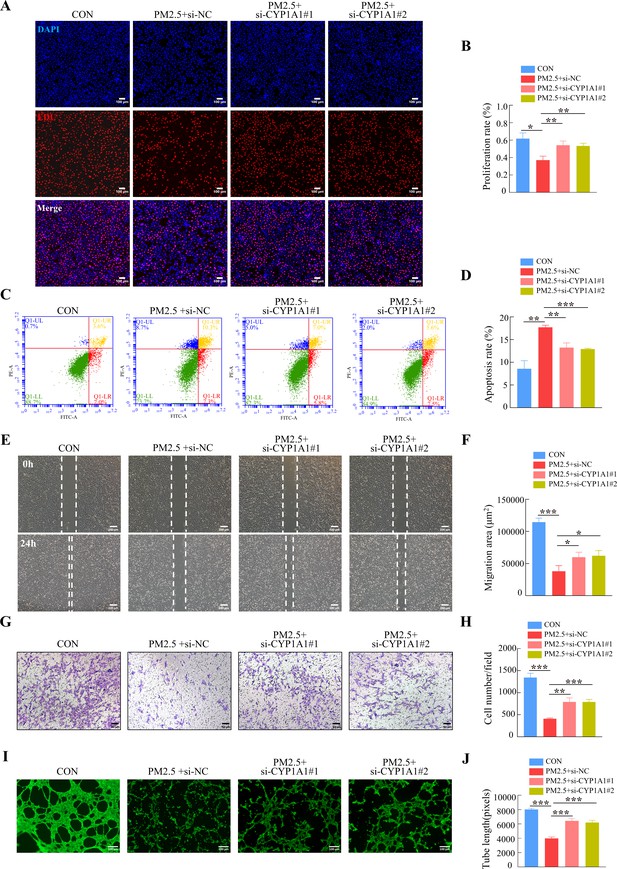
CYP1A1 knockdown reversed HTR8/SVneo cell dysfunction induced by PM2.5.
(A) Images showing the proliferation of HTR8/SVneo (PM2.5: 100 μg/mL). The nuclei were stained with DAPI in blue, the cells in the proliferation stage were stained with EDU in red. Scale bar, 100 μm. (B) The histogram analysis showed the proliferation rate of the cells in each group. (C) Detection of the percentage of apoptotic cells by flow cytometry (PM2.5: 100 μg/mL). (D) The histogram analysis indicated the apoptosis rate of the cells in each group. (E) Representative images of the wound healing assay of HTR8/SVneo cells at the 0 and 24 hr time points (PM2.5: 100 μg/mL). Scale bar, 200 μm. (F) The histogram showed the migration area (μm2) in each group. (G) Representative images of the Transwell invasion assay after 24 hr in the different groups. Scale bar, 50 μm. (H) The histogram indicated the cell counts/field in each group. (I) Representative images showing cell tube formation after 4 hr. Scale bar, 100 μm. (J) Histogram showed the quantification of the tube length formed by HTR8/SVneo cells. (*, p<0.05; **, p<0.01; ***, p<0.001).

KLF9 bound to a specific region of the CYP1A1 promoter to drive transcriptional activity.
(A) HTR8/SVneo cells in the PM2.5-exposed or non-exposed groups were treated with 20 μM actinomycin D, and RNA was collected at different times for qRT-PCR to detect mRNA expression, and the non-linear fitted curves showed similar half-lives of CYP1A1 mRNA. (B) HTR8/SVneo cells were cotransfected with CYP1A1-pGL3-WT and pRL-TK (internal control), and then exposed to 100 µg/mL PM2.5 for 24 hr. Dual luciferase assays were used to detect the fluorescence intensity. (C and D) Expression of CYP1A1 mRNA in KLF9 knockdown and over-expression HTR8/SVneo cells as detected by qRT-PCR. (E) Western blotting was used to detect the modulation of CYP1A1 protein expression by KLF9. (F) Different human CYP1A1 promoter-luciferase reporter gene structures together with pRL-TK were co-transfected in HTR8/SVneo cells stably over-expressing KLF9. Schematic graph on the left showed the binding sites of the CYP1A1 promoter predicted from the motif of KLF9 and mutant reporter genes constructed based on sites. The bars on the right showed the relative luciferase activity of different CYP1A1 promoters in KLF9 overexpressing cells (KLF9) compared to control cells (Vector) using dual-luciferase reporter gene assays. (G) Chromatin from KLF9 over-expression HTR8/SVneo cells was subjected to ChIP assay using KLF9 antibody or control IgG. PCR amplification with primers spanning the –96/+16 bp region of the CYP1A1 promoter was performed. A 2% agarose gel electrophoresis was performed on PCR products. (H) RT-qPCR analysis quantitatively demonstrated that KLF9 overexpression increased its binding to the endogenous CYP1A1 promoter. (I) Immunofluorescence staining showed co-staining of CYP1A1 (green) or KLF9 (green), with CK7 (red) and DAPI (blue) in human placental tissue. Scale bar, 500 μm. (J) Data shown were representative results of three repeated assays and are represented as mean ± SD. (si-KLF9#1, si-KLF9#2: siRNAs to knockdown KLF9 expression; si-NC: siRNAs Negative Control; * p<0.05, ** p<0.01, *** p<0.001).
-
Figure 9—source data 1
Labelled gel images.
(Figure 9E-KLF9) The expression of KLF9 expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-β-actin) The expression of β-actin expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-Vector-KLF9-KLF9) The expression of KLF9 expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9E-Vector-KLF9-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9E-Vector-KLF9-β-actin) The expression of β-actin expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9G-ChIP) Chromatin from KLF9 over-expression HTR8/SVneo cells was subjected to ChIP assay using KLF9 antibody or control IgG. PCR amplification with primers spanning the –96/+16 bp region of the CYP1A1 promoter was performed. A 2% agarose gel electrophoresis was performed on PCR products.
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig9-data1-v2.zip
-
Figure 9—source data 2
Raw unlabelled gel images.
(Figure 9E-KLF9) The expression of KLF9 expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-β-actin) The expression of β-actin expression in the HTR8/SVneo (si-NC, si- KLF9#1, si- KLF9#2). (Figure 9E-Vector-KLF9-KLF9) The expression of KLF9 expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9E-Vector-KLF9-CYP1A1) The expression of CYP1A1 expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9E-Vector-KLF9-β-actin) The expression of β-actin expression in the HTR8/SVneo (CON, KLF9 over expression). (Figure 9G-ChIP) Chromatin from KLF9 over-expression HTR8/SVneo cells was subjected to ChIP assay using KLF9 antibody or control IgG. PCR amplification with primers spanning the –96/+16 bp region of the CYP1A1 promoter was performed. A 2% agarose gel electrophoresis was performed on PCR products.
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig9-data2-v2.zip
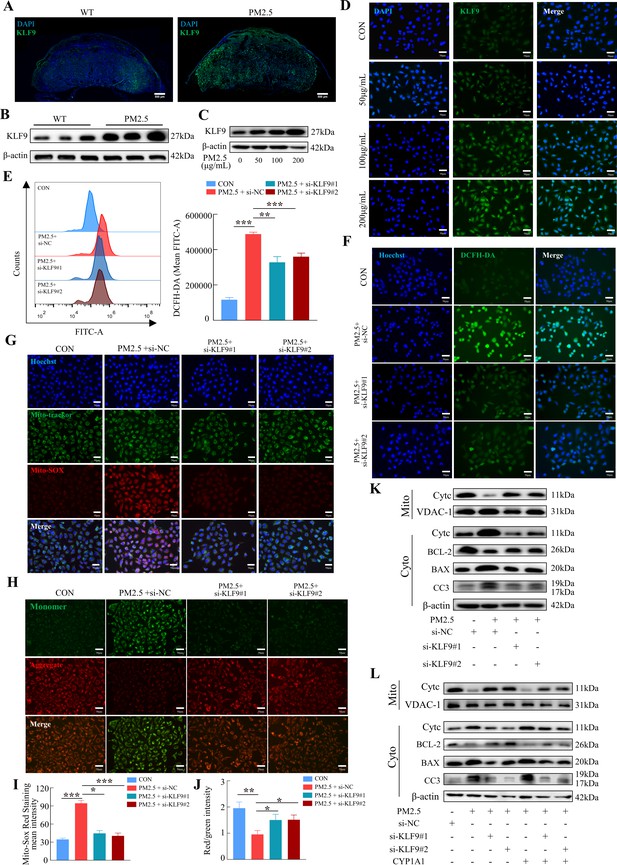
The KLF9/CYP1A1 signaling pathway was essential in PM2.5-induced oxidative stress damage and mitochondrial apoptosis in HTR8/SVneo cells.
(A) Expression of KLF9 in mice placental tissue sections was detected by immunofluorescence staining. The nuclei were stained with DAPI in blue. KLF9 was stained with a specific antibody (green). Scale bar, 500μm. (B) Detection of KLF9 expression in mouse placental tissues by western blotting (n=3). (C) The expression of KLF9 in HTR8/SVneo at different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL) were measured using western blotting. (D) Immunofluorescence images of KLF9 in HTR8/SVneo treated with different concentrations of PM2.5 (50 μg/mL, 100 μg/mL, 200 μg/mL). The nuclei were stained with DAPI (blue), and KLF9 was stained with specific antibody (green). Scale bar, 50 μm. (E) Quantitative detection of intracellular ROS by flow cytometry after staining with DCFH-DA. The histogram showed the mean FITC in each group. (F) Representative images of DCFH-DA staining for intracellular ROS (PM2.5: 100 μg/mL). The nuclei were stained with Hoechst (blue), the ROS in cells were stained with DCFH-DA (green). Scale bar, 50 μm. (G) Mitochondrial ROS levels in HTR8/SVneo (PM2.5: 100 μg/mL). Hoechst: labelling the nuclei of cells in blue; Mito-tracker: labelling the mitochondrial sites; Mito-sox: labelling the mitochondrial ROS. Scale bar, 50 μm. (H) Mitochondrial membrane potential was assessed based on JC-1 staining (PM2.5: 100 μg/mL). Scale bar, 50 μm. (I) The histogram indicated the Mito-SOX Red Staining mean intensity in each group. (J) The histogram indicated the Red/Green intensity in each group. (K) Mitochondrial apoptosis-associated protein expression levels in the mitochondria and cytoplasm were assessed using western blotting after knockdown of KLF9 (PM2.5: 100 μg/mL). Cyt-C expression was detected in mitochondria and cytoplasm, and BCL-2, BAX and Cleaved-caspase 3 (CC3) expression levels in the cytoplasm were detected. The mitochondrial marker VDAC and the cytosol marker β-actin were used to identify the purity of mitochondria in the extract. (L) Detection of mitochondrial apoptosis-associated protein expression in mitochondria and cytoplasm after KLF9 knockdown and CYP1A1 overexpression using western blotting.
-
Figure 10—source data 1
Labelled gel images.
(Figure 10B-KLF9) The expression of KLF9 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 10B-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 10C-KLF9) The expression of KLF9 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 10C-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 10K-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2) (Figure 10L-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression). (Figure 10L-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1 A1 overexpression, PM2.5+si-KLF9#1+CYP1 A1 overexpression, PM2.5+si-KLF9#2+CYP1 A1 overexpression).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig10-data1-v2.zip
-
Figure 10—source data 2
Raw unlabelled gel images.
(Figure 10B-KLF9) The expression of KLF9 expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 10B-β-actin) The expression of β-actin expression in the placental tissue of Wild Type (The first three columns) and PM2.5-treated mice (the last three columns). (Figure 10C-KLF9) The expression of KLF9 expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50μg/mL, 100μg/mL, 200μg/mL). (Figure 10C-β-actin) The expression of β-actin expression in the PM2.5-treated HTR8/SVneo cells (PM2.5 concentration: 0. 50μg/mL, 100μg/mL, 200μg/mL). (Figure 10K-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2). (Figure 10K-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2). (Figure 10K-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si- KLF9#1, PM2.5+si- KLF9#2) (Figure 10L-Mito-Cytc) The expression of Cytc expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Mito-VDAC-1) The expression of VDAC-1 expression in mitochondria of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Cyto-Cytc) The expression of Cytc expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Cyto-BCL-2) The expression of BCL-2 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Cyto-BAX) The expression of BAX expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Cyto-Cleaved-Caspase3) The expression of Cleaved-Caspase3 expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression). (Figure 10L-Cyto-β-actin) The expression of β-actin expression in cytoplasm of HTR8/SVneo (si-NC, PM2.5+si-NC, PM2.5+si-KLF9#1, PM2.5+si-KLF9#2, PM2.5+CYP1A1 overexpression, PM2.5+si-KLF9#1+ CYP1A1 overexpression, PM2.5+si-KLF9#2+ CYP1A1 overexpression).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig10-data2-v2.zip
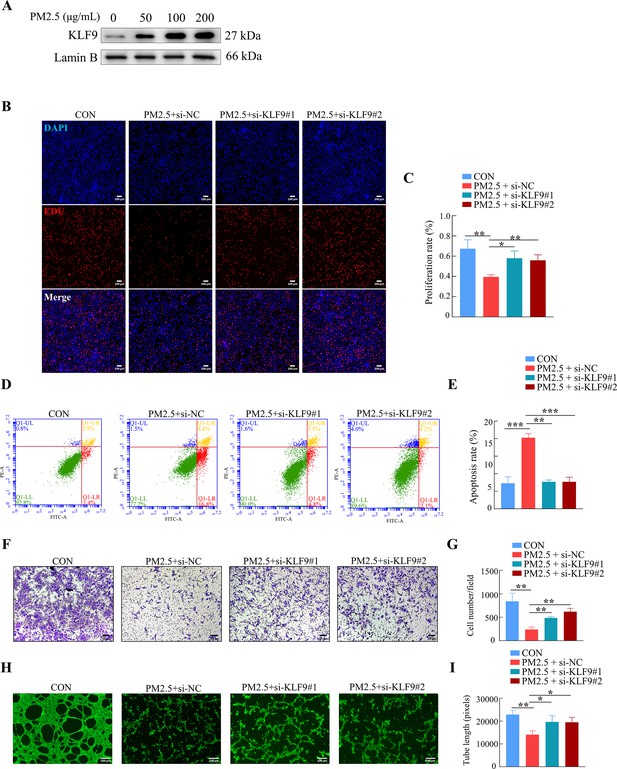
KLF9 was involved in PM2.5-induced dysfunction in HTR8/SVneo cells.
(A) Images showing the proliferation of HTR8/SVneo (PM2.5: 100 μg/mL). The nuclei were stained with DAPI in blue, the cells in the proliferation stage were stained with EDU in red. Scale bar, 100 μm. (C) Histogram analysis indicated the proliferation rate of the cells in each group. (D) Detection of the percentage of apoptotic cells by flow cytometry assay (PM2.5: 100 μg/mL). (E) Histogram analysis indicated the apoptosis rate of the cells in each group. (F) Representative images of the Transwell invasion assay after 24 hr in the different groups. Scale bar, 50 μm. (G) The histogram indicated the cell counts/field in each group. (H) Representative images showed the cell tube formation at 4 hr. Scale bar, 100 μm. (I) Histogram showed the quantification of the tube length of HTR8/SVneo cells. (*, p<0.05; **, p<0.01; ***, p<0.001).
-
Figure 10—figure supplement 1—source data 1
Labelled gel images.
(Figure 10—figure supplement 1A–KLF9) The expression of KLF9 expression in the HTR8/SVneo cell nucleus after treated with PM2.5 (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 10—figure supplement 1B–Lamin B) The expression of Lamin B expression in the HTR8/SVneo cell nucleus after treated with PM2.5 (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig10-figsupp1-data1-v2.zip
-
Figure 10—figure supplement 1—source data 2
Raw unlabelled gel images.
(Figure 10—figure supplement 1A–KLF9) The expression of KLF9 expression in the HTR8/SVneo cell nucleus after treated with PM2.5 (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL). (Figure 10—figure supplement 1B–Lamin B) The expression of Lamin B expression in the HTR8/SVneo cell nucleus after treated with PM2.5 (PM2.5 concentration: 0. 50 μg/mL, 100 μg/mL, 200 μg/mL).
- https://cdn.elifesciences.org/articles/85944/elife-85944-fig10-figsupp1-data2-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CYP1A1 | GenBank | HGNC:HGNC:2595 | |
| Gene (Homo sapiens) | KLF9 | GenBank | HGNC:HGNC:1123 | |
| Antibody | anti-CYP1A1 (Rabbit polyclonal) | Proteintech | Cat#13241–1-AP | WB (1:1000) IHC (1:500) |
| Antibody | anti-KLF9 (Rabbit polyclonal) | Abcam | Cat#ab227920 | WB (1:1000) IHC (1:500) ChIP (1:1000) |
| Antibody | anti-HO-1 (Rabbit polyclonal) | Cohesion Biosciences | Cat#CQA2561 | WB (1:1000) |
| Antibody | anti-NQO-1 (Rabbit polyclonal) | Cohesion Biosciences | Cat#CPA1342 | WB (1:1000) |
| Antibody | anti-GCLC (Rabbit polyclonal) | Cohesion Biosciences | Cat#CPA2092 | WB (1:1000) |
| Antibody | anti-SOD-1 (Rabbit polyclonal) | Cohesion Biosciences | Cat#CPA1476 | WB (1:1000) |
| Antibody | anti-CK-7 (Rabbit monoclonal) | Abcam | Cat#ab68459 | IHC (1:500) |
| Antibody | anti-β-actin (Mouse monoclonal) | Proteintech | Cat#66009 | WB (1:1000) |
| Antibody | anti-Lamin B | Proteintech | Cat#12987–1-AP | WB (1:1000) |
| Cell line (Homo sapiens) | HTR8-SVneo (Homo sapiens) | ATCC | NO.CRL-3271 | |
| Sequence-based reagent | siCYP1A1#1_F | This paper | siRNA sequence | GGUAUGUGGUGGUAUCAGUTT |
| Sequence-based reagent | siCYP1A1#1_R | This paper | siRNA sequence | ACUGAUACCACCACAUACCTT |
| Sequence-based reagent | siCYP1A1#2_F | This paper | siRNA sequence | CCUUCAAGGACCUGAAUGATT |
| Sequence-based reagent | siCYP1A1#2_R | This paper | siRNA sequence | UCAUUCAGGUCCUUGAAGGTT |
| Sequence-based reagent | siKLF9#1_F | This paper | siRNA sequence | GCCCAUUACAGAGUGCAUATT |
| Sequence-based reagent | siKLF9#1_R | This paper | siRNA sequence | UAUGCACUCUGUAAUGGGCTT |
| Sequence-based reagent | siKLF9#2_F | This paper | siRNA sequence | GGAGUGACCACCUCACAAATT |
| Sequence-based reagent | siKLF9#2_R | This paper | siRNA sequence | UUUGUGAGGUGGUCACUCCTT |
| Sequence-based reagent | CYP1A1_F | This paper | PCR primers | TGGCATCCTCTACAGACTCCTG |
| Sequence-based reagent | CYP1A1_R | This paper | PCR primers | CTTCAGGTTGCGTGCCATCTCA |
| Sequence-based reagent | KLF9_F | This paper | PCR primers | CTACAGTGGCTGTGGGAAAGTC |
| Sequence-based reagent | KLF9_R | This paper | PCR primers | CTCGTCTGAGCGGGAGAACTTT |
| Sequence-based reagent | CYP1A1-promoter_F | This paper | PCR primers | CTGCTTCTCCCTCCATCT |
| Sequence-based reagent | CYP1A1-promoter _R | This paper | PCR primers | GGAACTGTCACCTTCAGG |
| Commercial assay or kit | GSH | MilliporeSigma | MAK440 | |
| Commercial assay or kit | SOD | Beyotime Institute of Biotechnology, China | S0103 | |
| Commercial assay or kit | MDA | Beyotime Institute of Biotechnology, China | S0131S | |
| Commercial assay or kit | EDU | Beyotime Institute of Biotechnology, China | C0075 | |
| Commercial assay or kit | Apoptosis kit | BD Biosciences | 559763 | |
| Commercial assay or kit | TUNEL staining | Elabscience Biotechnology | E-CK-A320 | |
| Commercial assay or kit | ChIP assay kit | Beyotime Institute of Biotechnology | P2078 | |
| Commercial assay or kit | Dual luciferase reporter assay kit | Promega Corporation | E1910 | |
| Recombinant DNA reagent | pLVX-Puro (plasmid) | purchased from Tsingke biotechnology | plasmid sequences were verified by Tsingke biotechnology | |
| Recombinant DNA reagent | pLVX-CYP1A1- 3×FLAG (plasmid) | purchased from Tsingke biotechnology | plasmid sequences were verified by Tsingke biotechnology | |
| Recombinant DNA reagent | pLVX-KLF9- 6×His (plasmid) | purchased from Tsingke biotechnology | plasmid sequences were verified by Tsingke biotechnology | |
| Chemical compound, drug | NAC | MillporeSigma | A7250 | |
| Chemical compound, drug | JC-1 | Beyotime Institute of Biotechnology | C2005 | |
| Chemical compound, drug | Actinomycin D | Beyotime Institute of Biotechnology | GC16866 | |
| Software, algorithm | Image J | National Institutes of Health | V 1.8.0 | |
| Software, algorithm | GraphPad | GraphPad Software | V 8.0 |
Additional files
-
Supplementary file 1
The gestation and characteristics of the pregnant women (n=31).
- https://cdn.elifesciences.org/articles/85944/elife-85944-supp1-v2.docx
-
Supplementary file 2
The sequence of siRNAs used in this study.
- https://cdn.elifesciences.org/articles/85944/elife-85944-supp2-v2.docx
-
Supplementary file 3
The sequence of primers used in this study.
- https://cdn.elifesciences.org/articles/85944/elife-85944-supp3-v2.docx
-
Supplementary file 4
The information of antibodies used in this study.
- https://cdn.elifesciences.org/articles/85944/elife-85944-supp4-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85944/elife-85944-mdarchecklist1-v2.pdf