Endoparasitoid lifestyle promotes endogenization and domestication of dsDNA viruses
Figures
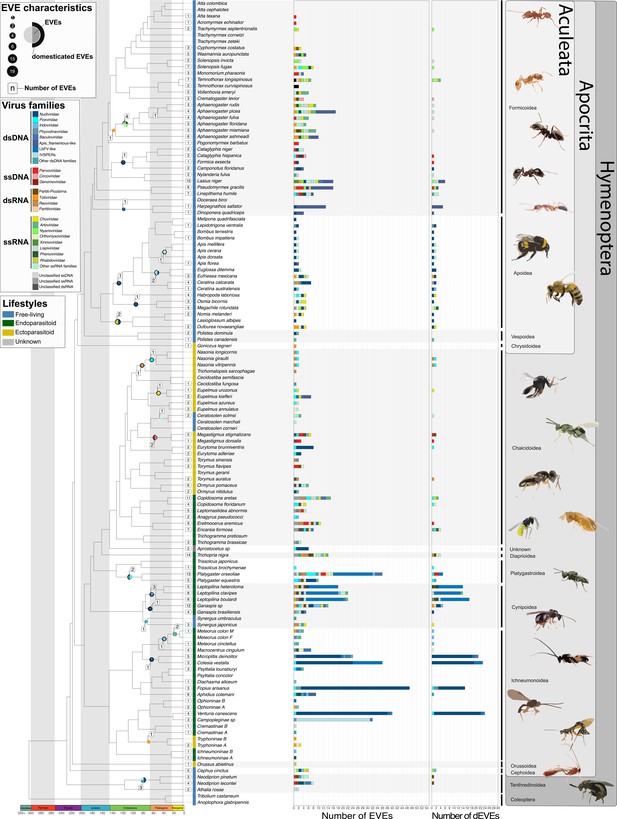
Endogenous Viral Elements (EVEs ) and their domestication status in Hymenoptera.
Lifestyles are displayed next to species names (blue: free-living, green: endoparasitoid, yellow: ectoparasitoid, gray: unknown). The number of EVEs and domesticated EVEs (dEVEs) found in each species are represented respectively by the first and second facets of the horizontal histograms. Colors along these histograms indicate the potential donor viral families (where blue tones correspond to viral double-stranded DNA (dsDNA) viruses, red tones to single-stranded DNA (ssDNA) viruses, orange/brown tones to dsRNA viruses, and green tones to ssRNA genomes). EVEs shared by multiple species and classified within the same event are represented by circles whose size is proportional to their number; those that are considered as dEVEs are surrounded by a black border. Numbers in the white boxes correspond to the number of endogenization events inferred. As an example, Megastigmus dorsalis and Megastigmus stigmatizans are ectoparasitoids (yellow) sharing a common endogenization event (within the Cluster21304, see Figure 1—figure supplement 1) that likely originated from an unclassified dsRNA virus (gray color in circle), and shows no sign of domestication (no black border around the gray part of the circle). The figure was inspired by the work of Peters et al., 2017. Details on the phylogenetic inference and time calibration can be found in the Material an methods section; bootstrap information can be found in Figure 1—figure supplement 7; details on lifestyle assignation can be found in Supplementary file 2. All Cluster sequence alignments from loci scored from A to X can be found within the Figure 1—source data 1.
-
Figure 1—source data 1
File including all aligned cluster sequences scored from A to X.
- https://cdn.elifesciences.org/articles/85993/elife-85993-fig1-data1-v1.zip
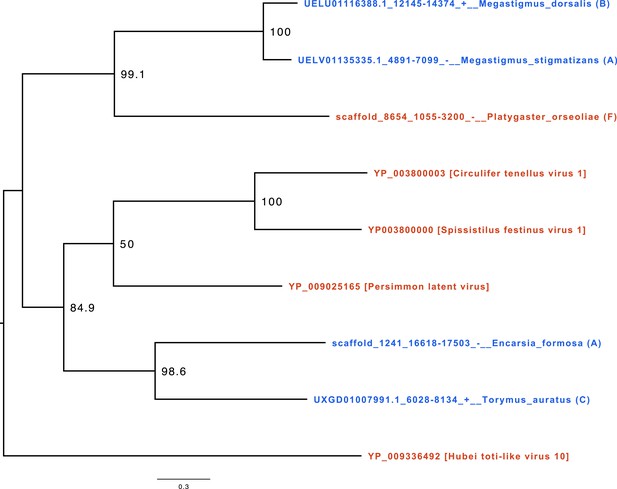
Example of endogenization events.
The phylogeny of cluster21304 corresponds to the clustering of a set of viral and candidate viral insertion genes sharing a homology. In red are represented the loci of viral origin, and in blue are represented the loci probably endogenized endogenous viral elements (EVEs). The letter at the end of the taxon label represents the endogenization score assigned to the candidate (see details in Materials and methods). In this example, we found two singular endogenization events in the species endoparasitoid Encarsia formosa (annotated A and thus presenting a depth of coverage non-significantly different from the distribution of the BUSOs of the genome as well as at least one transposable element and/or one eukaryotic gene) and ectoparasitoid Torymus auratus (annotated C and thus presenting only a depth of coverage non-significantly different from the distribution of the BUSOs of the genome). Since these two species do not share a close common ancestor in the phylogeny and come from two different families, the algorithm, therefore, assigned them to two independent viral endogenization events. The viral locus found in the assembly of the endoparasitoid species Platygaster orseoliae was annotated F, meaning that the depth of coverage deviated significantly from the BUSCO distribution of the genome and that no TEs and less than five eukaryotic genes were found in the scaffold containing the candidate insertion. Finally, the two loci belonging to the ectoparasitoid species Megastigmus stigmantizans and Megastigmus dorsalis both show a score supporting viral endogenization. Furthermore, these species exhibit a doubly monophyletic clade (high bootstrap score) within the gene phylogeny and within the species phylogeny, suggesting that they acquired this viral gene from their closest common ancestor about 20 million years ago. All newick phylogenies are available in Supplementary file 12.
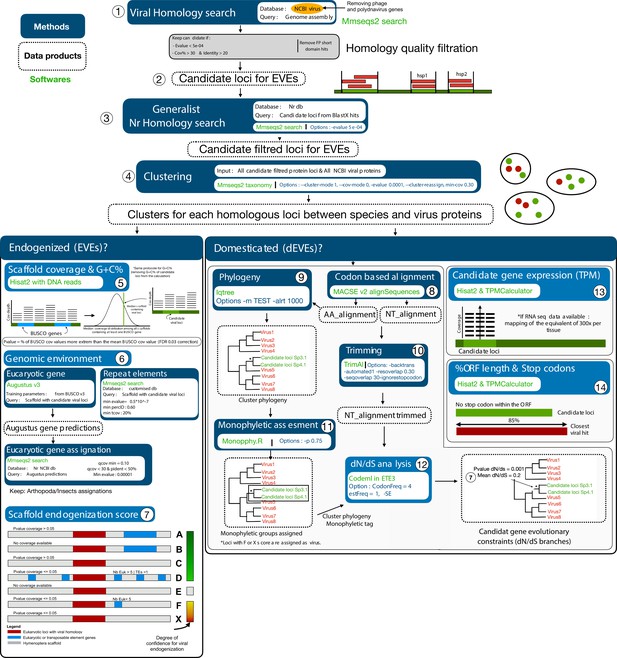
Simplified summary of the bioinformatics pipeline for the detection and validation of candidates for endogenization and domestication.
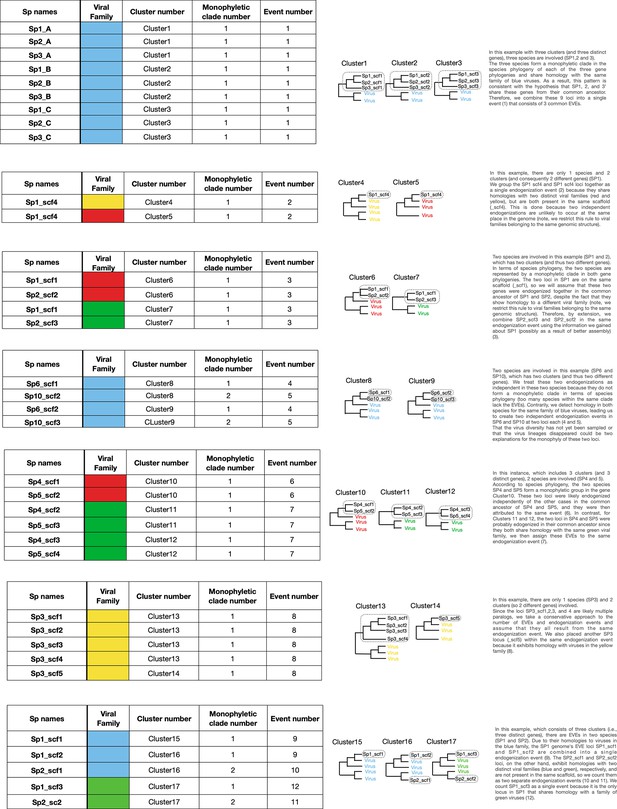
Canonical examples of endogenization events inferred by our pipeline.
The column ‘Sp names’ contains the species name, followed by the name of the scaffold in which the endogenous viral element (EVE) has been identified. The ‘Viral family’ column refers to the putative viral family that donated the EVEs. The column ‘Cluster number’ corresponds to the number of the cor number of the monophyletic clade within a cluster (can be a single locus or multiple loci). The column ‘Event number’ is the number given to single/multiple EVEs that derive from the same endogenization event.
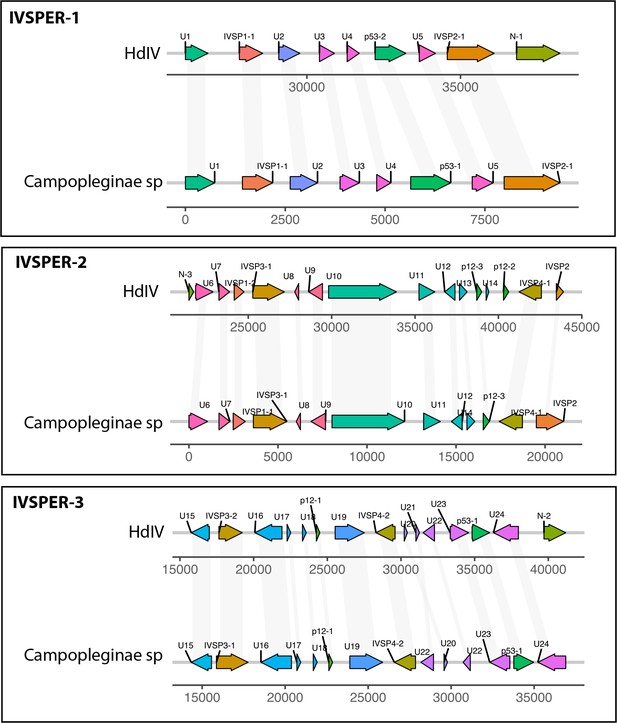
IVSPER genes identified in the Campopleginae genome.
The figure compares the synteny of the IVSPER between Hyposoter didymator ichnovirus (HdIV) and the Campoplegninae of our dataset. Homologous genes with synteny between the two species are indicated by gray shading. The direction of the arrows corresponds to the sense and anti-sense strands. The color of the boxes is unique to each IVSPER.
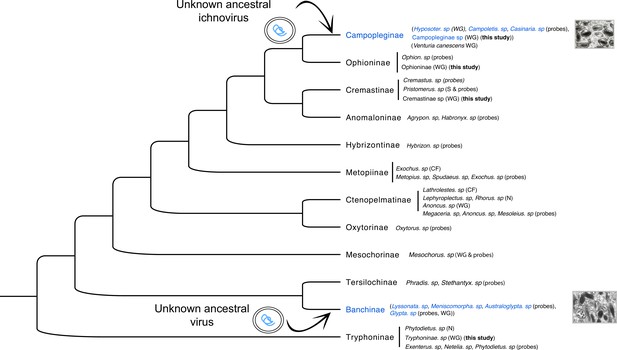
Cladogram of the Ophioniformes group, illustrating the two independent endogenization events of two unknown viruses in Banchinae and Campopleginae lineages.
The phylogeny includes 12 subfamilies of the Ophioniformes group within the superfamily Ichneumonoidea. Several species of these subfamilies have been examined for the presence of ichnovirus-like polydnaviruses: by negative staining of calyx fluid (N), TEM of ovarian sections (S), visual examination of the calyx fluid (CF), probes from ichnovirus replication or structural proteins (probes) or by IVSPER sequence homology on whole genome assemblies (WG). The subfamilies and species in blue correspond to those positive for a double-stranded DNA (dsDNA) virus endogenization from an unknown origin (ichnovirus-like). The others (in black) were negative for endogenized ichnovirus-like elements. The phylogeny is inspired from Sharanowski et al., 2021, and the data reported comes from Sharanowski et al., 2021; Béliveau et al., 2015; Cusson, 2012; Legeai et al., 2020.
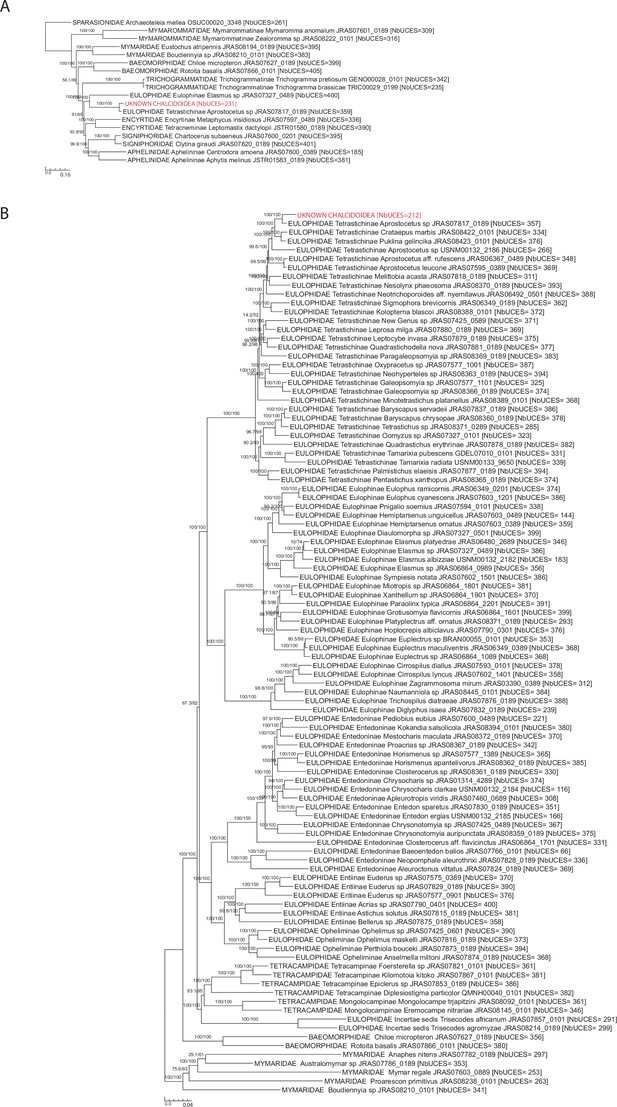
Ultra conserved element (UCE) trees built to assign to species the unknown Chalcidoidea sequenced with the pool of P. orseoliae.
(A) Phylogeny of early diverging families of Chalcidoidea (423 UCEs and 127,979 bp were analyzed to get the tree, best-fit model = GTR + F + R10). (B) Phylogeny of the family Eulophidae to which the unknown sample was inferred to belong to (408 UCEs and 77,514 bp were analyzed to get the tree, best-fit model = GTR + F + R10). For both trees, SH-aLRT/UFBoot are shown at nodes; the number of UCEs analyzed for each sample is indicated between brackets and the unknown sample is highlighted in red.
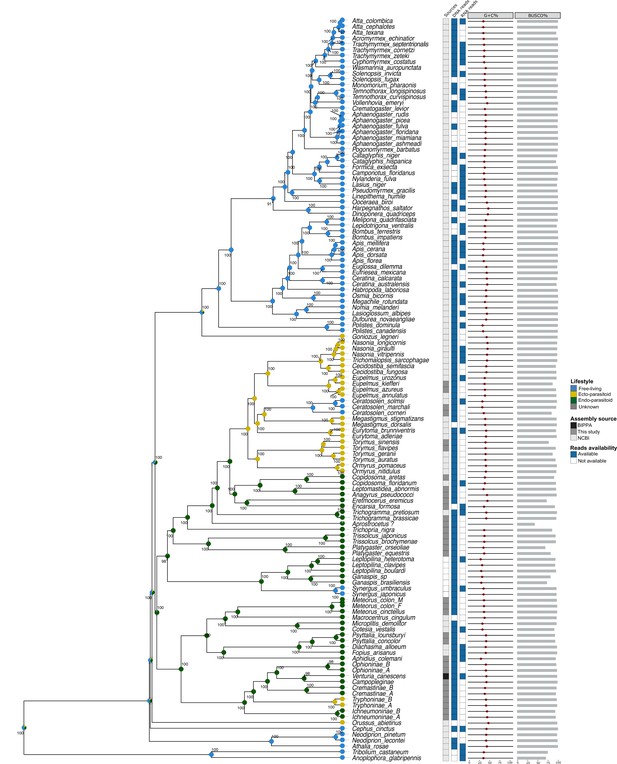
Source of the datasets and availability of the reads.
Phylogeny of 124 Hymenoptera species. Two Coleoptera species were used to root the tree. The aLRT bootstrap scores are represented along the nodes. The sources refer to the platform or laboratory in which the assemblages come from (This study, BIPPA: BioInformatics Platform for Agroecosystem Arthropods, NCBI: National Center for Biotechnology Information). The assemblies for which raw DNAseq or RNAseqs reads were available are listed in the column DNA or RNA reads. The G+C% column reflects the average G+C rate for each assembly, and the BUSCO% column reflects the rate of complete or partial BUSCOs found via the analysis with BUSCO V3. Posterior Bayesian lifestyle inference distribution for each node and tips are represented by colored pie charts.
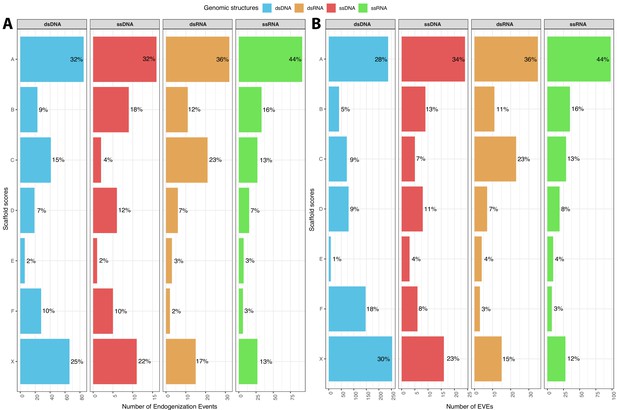
Representation of the score distribution among different virus genome types.
A represents the distribution of the number of endogenization Events and B the number of endogenous viral elements (EVEs). The percentage of Events or EVEs is shown next to the bars.
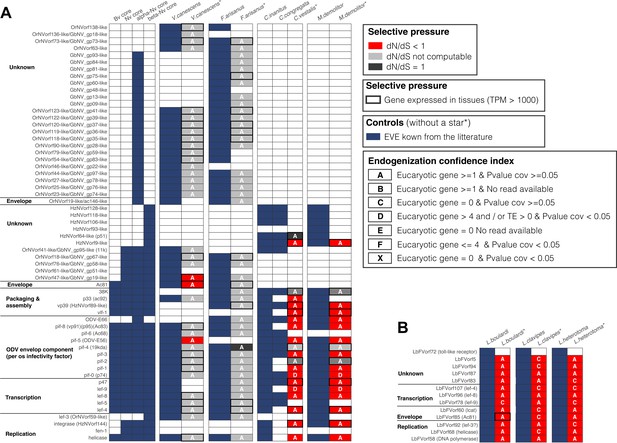
Heatmap representing the viral genes known to be domesticated by Hymenoptera.
The panel (A) refers to the four known cases (Venturia canescens, Fopius arisanus, Cotesia congregata, and Microplitis demolitor) involving Nudivirus donors while the panel (B) refers to the known case involving LbFV donors in three Leptopilina species. Complete parasitoid wasp genomes information was available for Microplitis demolitor, Venturia canescens, Fopius arisanus, and Cotesia congregata, while only partial genomic data were available for Chelonus inanitus. Each row indicates a gene that has been identified previously as being endogenized in at least one species. In (A), the first four columns indicate whether the gene is a core gene for baculoviruses (Bv), Nudiviruses (Nd), alpha-nudiviruses (alpha-Nv), or beta-nudivirus (beta-nv). The following columns indicate the presence of each gene based on the literature (in blue) and based on our pipeline (columns with a star symbol). The colors indicate the inferred selection pressure acting on each gene (dN/dS) and the letters A, B, C, D, E, and X represent the degree of confidence in the endogenization. The box is framed in black if our pipeline detected expression (TPM>1000).
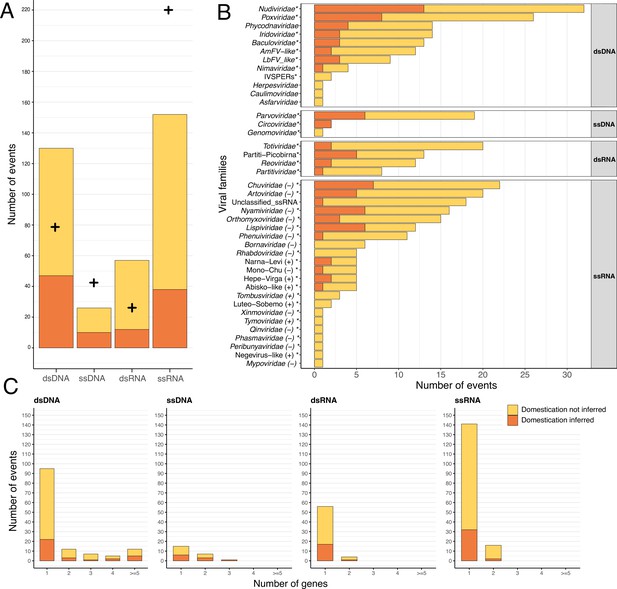
Endogenization involves all types of viral genomic structures.
In all three panels, events inferred as corresponding to domestications are displayed in orange, while events not inferred as domestications are displayed in yellow. (A) Distribution of the number of events inferred, according to the four categories of viral genomic structures. The crosses refer to the expected number of endogenization events for each category based on its estimated relative abundance in insects (see details in Materials and methods and virus-infecting data in Supplementary file 5). The data used in this figure can be found in Supplementary file 6 in the ‘Figure_data’ sheet. (B) Distribution of the various viral families involved in endogenization events. The polarity of single-stranded RNA (ssRNA) viruses is displayed next to the family name. Events involving multiple putative families (i.e. where several viral families are present in the same scaffold) have been excluded from the count. The star next to the family name indicates that the viral family is known to infect insects. (C) Distribution of the number of endogenous viral elements (EVEs) per event across viral categories.
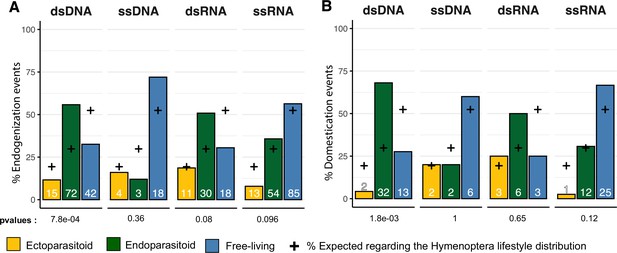
Endogenization and domestication of double-stranded DNA (dsDNA) viruses are most prevalent in endoparasitoid species.
(A) Distribution of viral endogenization events (Event) and B of domestication events (dEVEs) across Hymenoptera lifestyles. Crosses indicate the expected proportion of events associated with the different lifestyles, based on the respective frequencies in our database (ectoparasitoid = 24/124, endoparasitoid = 37/124, free-living = 63/124). The p-values are the results of Fisher’s tests comparing the observed and expected distributions. Numbers inside the bars indicate the absolute numbers of events inferred. The ancestral states of the nodes, in terms of lifestyle, were inferred in a Bayesian analysis (see details in Materials and methods). The data used in this figure can be found in Supplementary file 6.
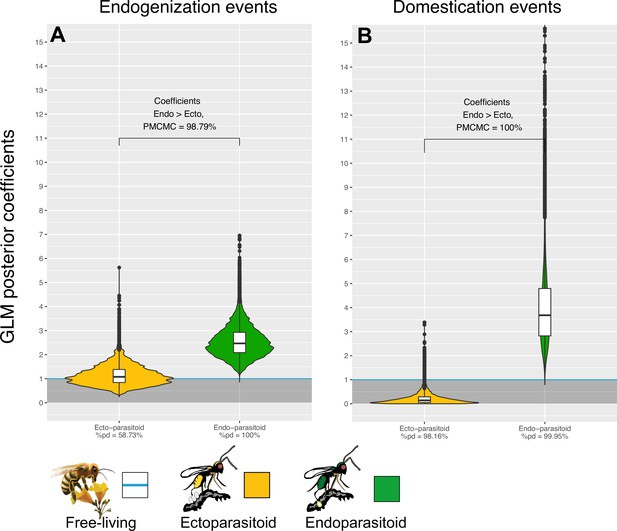
Endogenization and domestication of double-stranded DNA (dsDNA) viruses are more frequent in endoparasitoid species.
Violin plots represent the posterior distribution of the coefficients obtained under the different GLM models (after exponential transformation to obtain a rate relative to free-living species). The coefficients are derived from 1000 independent GLM models, where 1000 probable scenarios of ancestral states at nodes were sampled randomly among the MCMCM iterations (see details in Materials and methods). Branches from nodes older than 160 million years were removed from the dataset. The %pd is the probability of direction and indicates the proportion of the posterior distribution where the coefficients have the same sign as the median coefficient. indicates the proportion of MCMC iterations where the coefficient obtained for endoparasitoid species is higher than for ectoparasitoid species. All statistical summaries of the Bayesian GLM models can be found in Supplementary file 6.
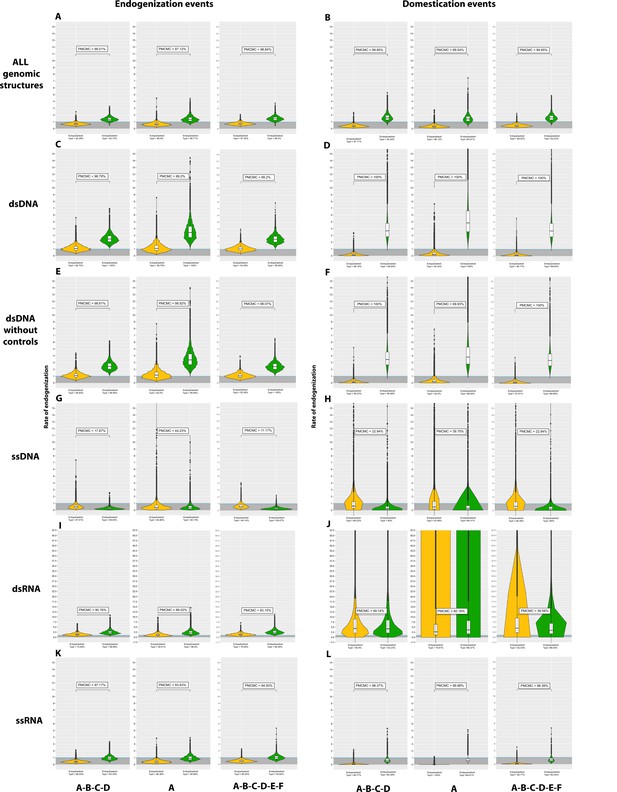
Violin plots of the posterior distribution of GLM coefficients in relation to Hymenoptera lifestyle.
The ectoparasitoid lifestyle is in yellow, the endoparasitoid lifestyle is in green, and the free-living lifestyle is in blue. A binomial negative zero-inflated GLM model was used, with free-living as the intercept. The distribution of the coefficients is given after exponential transformation. The Y-axis thus corresponds to the multiplicative factor of the number of endogenization and/or domestication of endogenous viral elements (EVEs) and/or events relative to free-living species. The coefficients are derived from 1000 GLM models adjusted on 1000 randomly selected probable scenarios (>90 CI) of ancestral states at nodes. Branches from nodes older than 160 million years have been removed from the dataset. The ROPE% is the percentage of the posterior distribution of coefficients below the intercept. There was no significant interaction between lifestyles and branch size, and the factor ‘branch size’ was, therefore, added to the model as an additive effect.
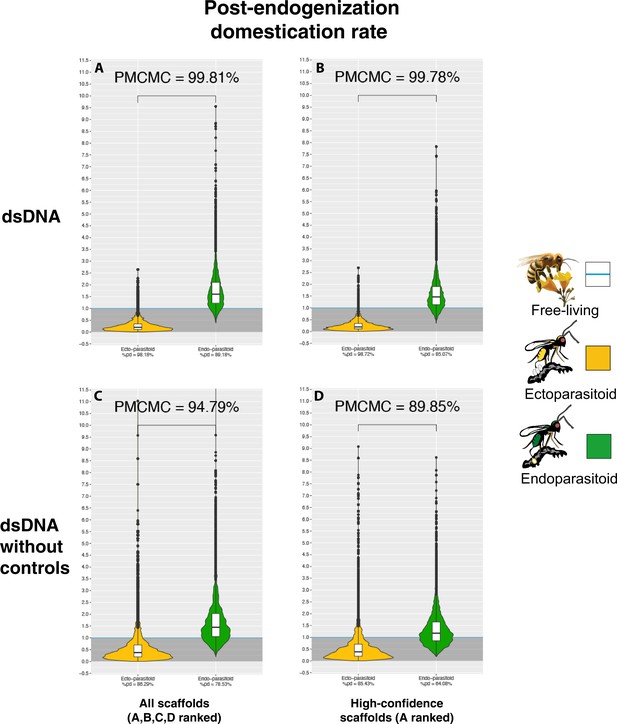
Violin plots of the posterior distribution of dEvents GLM coefficients in relation to wasp lifestyle (corrected for Events rates).
The ectoparasitoid lifestyle is in yellow, the endoparasitoid lifestyle is in green, and the free-living lifestyle is in blue (the intercept). Coefficients have been transformed into exponential and correspond to the posterior distribution of the coefficients of a binomial logistic regression GLM model, where the lifestyle free-living stands for the intercept. The Y-axis corresponds to the multiplicative factor of the number of dEvents (corrected for Events rates) correlative to free-living species. The coefficients are derived from 1000 GLM models adjusted on 1000 randomly selected probable scenarios (>90 CI) of ancestral states at nodes. Branches from nodes older than 160 million years have been removed from the dataset. The ROPE% is the percentage of the posterior distribution of coefficients below the intercept. The posterior distribution of the interaction coefficients between lifestyles and branch size was not informative, and the branch size factor was, therefore, added as an additive effect to the model. A- The corrected coefficient within all dEvents, B- The corrected coefficient within all dEvents without the control genomes, C- The corrected coefficient within all dEvents present in a scaffold annotated with a score A, D- The corrected coefficient within all dEvents present in a scaffold annotated with a score A and without the control genomes.
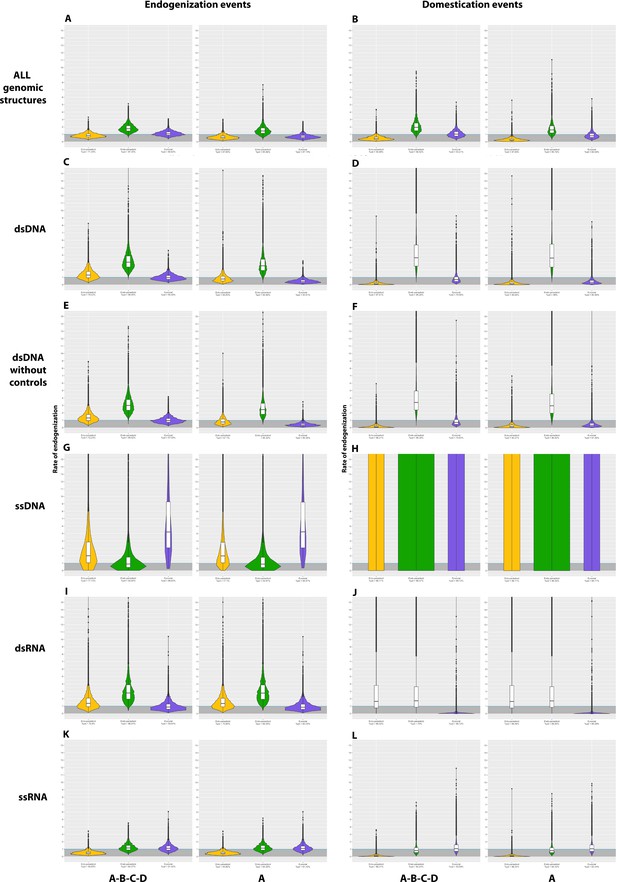
Violin plots of the posterior distribution of GLM coefficients in relation to Hymenoptera lifestyle.
The ectoparasitoid lifestyle is in yellow, the endoparasitoid lifestyle is in green, the free-living lifestyle is in blu.e and the eusocial lifestyle is in purple. A binomial negative zero-inflated GLM model was used, with free-living as the intercept. The distribution of the coefficients is given after exponential transformation. The Y-axis thus corresponds to the multiplicative factor of the number of endogenization and/or domestication of endogenous viral elements (EVEs) and/or events relative to free-living species. The coefficients are derived from 1000 GLM models adjusted on 1000 randomly selected probable scenarios (>90 CI) of ancestral states at nodes. Branches from nodes older than 160 million years have been removed from the dataset. The ROPE% is the percentage of the posterior distribution of coefficients below the intercept. There was no significant interaction between lifestyles and branch size, and the factor ’branch size’ was, therefore, added to the model as an additive effect.
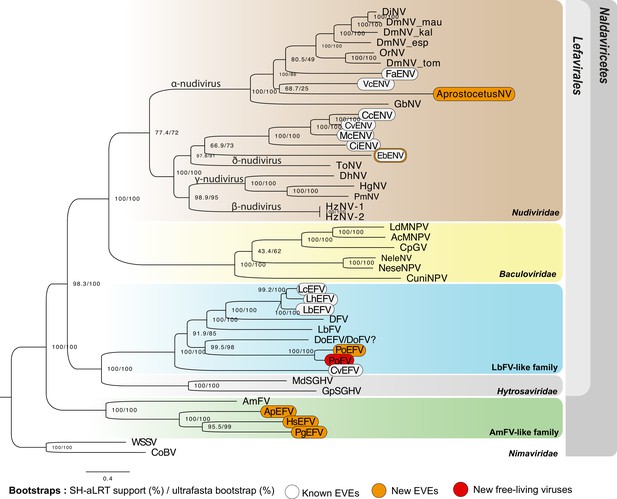
Phylogenetic relationships among endogenized and ‘free-living’ double-stranded DNA (dsDNA) viruses.
Specifically, this figure shows the relationships between Naldaviricetes double-stranded DNA viruses and endogenous viral elements (EVEs) from hymenopteran species, where at least three endogenization events were found. This tree was computed using maximum likelihood in Iqtree (v2) from a 38,293-long protein alignment based on the concatenation of 142 viral genes. Confidence scores (aLRT%/ultra-bootstrap support%) are shown at each node. The scale bar indicates the average number of amino acid substitutions per site. Previously known EVEs are in white, those from the present study in orange, and leaves inferred as free-living viruses are in red. All the best partitioned models as well as the number of genes used for each taxa can be found in Supplementary file 7. All free-living dsDNA viruses used in this phylogeny were obtained from published complete viral genomes. More details on the phylogenetic inference can be found in the methods. The ‘?’ for DoEFV/DoFV refers to the uncertainty regarding the endogenous/exogenous status of this sequence. All Cluster sequence alignments can be found in the Figure 5—source data 1.
-
Figure 5—source data 1
File including all aligned cluster sequences included in the Naldaviricetes phylogenetic analysis.
- https://cdn.elifesciences.org/articles/85993/elife-85993-fig5-data1-v1.zip
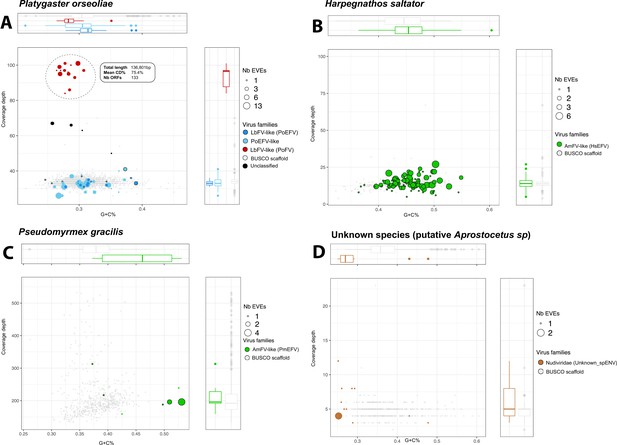
G+C% Coverage distribution of scaffold containing multiple endogenous viral elements (EVEs) Events.
The size of the dots corresponds to the number of candidate EVEs inside the scaffold. The color represents the genomic entity from which the EVE probably originated (brown: Nudiviridae, blue = LbFV, and green = AmFV). The red color refers to scaffolds showing free-living virus signatures. The gray color refers to scaffolds containing one or more BUSCO genes. The dots circled in black correspond to scaffolds that contain one or more eukaryotic genes and/or repeat elements.
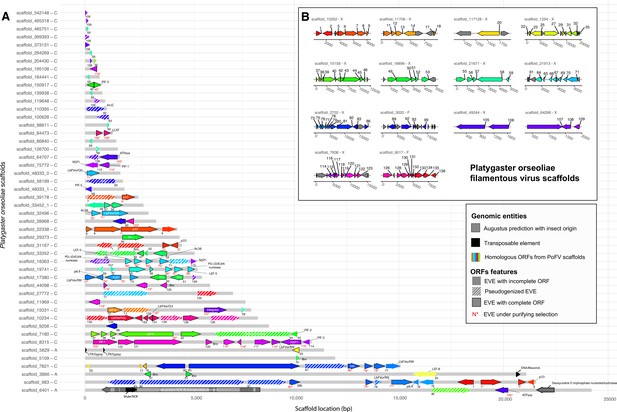
Genomic environment for the endogenous viral elements (EVEs) detected in Platygaster orseoliae.
The plot show regions homologous to viral ORFs in the Platygaster orseoliae filamentous virus (PoFV) genome (A). The colored regions correspond to the predicted ORFs in the PoFV genome (gray ORFS in PoFV scaffolds mean that no homologous EVEs were found in the genome of P.orseoliae.) (B), so the closer the colors, the closer the ORFs are in the PoFV genome. In gray are displayed the eukaryotic genes predicted by Augustus, with a dark color for exons, and light for introns. In black are displayed the transposable elements predicted by sequence homologies with the RepeatPeps protein database (max E-value = 5.866e-18). The letter followed by the scaffold name refers to the scoring given to the scaffold based on coverage and gene/ET presence information (see details in Materials and methods). The exact coordinates of the elements are referred to the x abscissa, which corresponds to the coordinates in base pairs and can be found in Supplementary file 11. Annotation is indicated in or next to the arrows. The number below each EVE corresponds to the homologous ORF number in the PoFV genome. Numbers are colored in red if the EVE has at least one paralog and if the computed dN/dS is below 1, suggesting purifying selection in the P.orseoliae genome. Arrow with black borders correspond to EVEs showing a complete ORF (>50% of the size of the best PoFV ORF). Hatched arrows correspond to pseudogenized EVEs (with premature stop codons). The color difference between black and white for the names of the proteins is for visual purposes only.
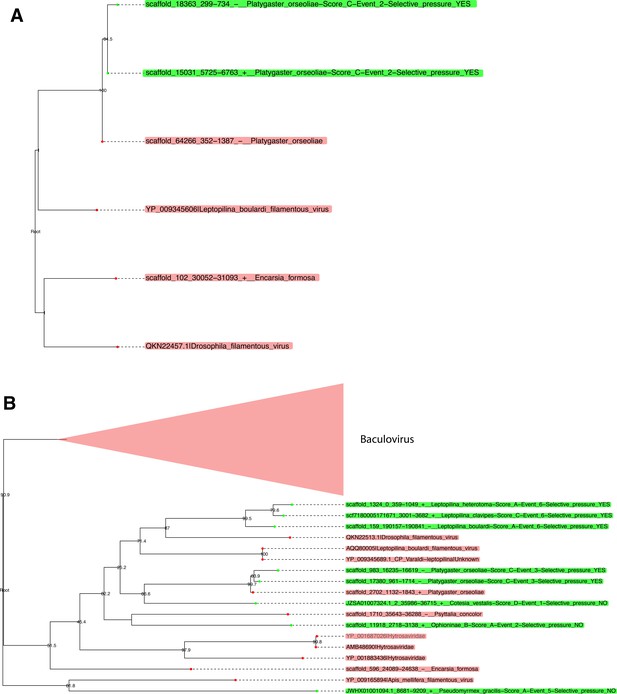
Phylogenies of LbFV-like proteins under purifying selection in Platygaster orseoliae genome.
The panel A represents the Cluster_25710 which corresponds to the integrase protein. The panel B represents the Cluster_26675 which corresponds to the ac81 protein. Taxa in red correspond to putative loci belonging to free-living viruses, while green taxa correspond to putative EVEs (the assigned family is indicated after the pipe). Green labels indicate the following information: (1) scaffold name, (2) start location, (3) end location, (4) strand, (5) Hymenoptera species, (6) endogenization index, (7) the inferred event number within the cluster phylogeny, and (8) whether the loci are found under purifying selection (YES) or not (NO).

Candidate endogenous viral elements (EVEs) in two ant species.
In gray are displayed the eukaryotic genes predicted by Augustus, with a dark color for exons, and light for introns. In black are displayed the transposable elements predicted by sequence homologies with the RepeatPeps protein database. The size of the scaffold is displayed below the name of each scaffold. The letter followed by the scaffold name refers to the scoring given to the scaffold based on coverage and gene/ET presence information (see details in Materials and methods). For the sake of representation, all scaffolds are represented at different scales, the exact coordinates are indicated in base pairs in the abscissa. Annotation is indicated above the arrows.
Additional files
-
Supplementary file 1
Summary statistics table for candidate EVEs.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp1-v1.xlsx
-
Supplementary file 2
General information regarding the species used in this study.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp2-v1.xlsx
-
Supplementary file 3
Summary statistics table for control endogenous viral elements (EVEs).
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp3-v1.xlsx
-
Supplementary file 4
Information for individual loci.
Endogenized loci (scoring from A to D) are displayed in the first sheet, whereas exogenous loci (from E to X) are displayed in the second sheet.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp4-v1.xlsx
-
Supplementary file 5
Table listing the names of virus species known to probably interact with insects.
The data is taken from the virushostdb database (Mihara et al., 2016) (version of 24/03/2023), which lists a wide variety of virus species associated with their presumed hosts. We have also added two important exploratory studies of RNA viruses (Shi et al., 2016; Wu et al., 2020). The viral genomic structures associated with the viruses were retrieved from the ICTV website (V2022_MSL38). Each column represents the information retrieved for each virus species from one of the three sources listed. The Hostdb_Host_lineage column corresponds to the information of the insect host observed interacting with the virus of interest. If a column with the suffix ’Shi’ or ’Haoming’ contains information for a virus species, then this means that this RNA virus species was found in their dataset in an insect.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp5-v1.xlsx
-
Supplementary file 6
Datasets and detailed statistics are presented in the manuscript.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp6-v1.xlsx
-
Supplementary file 7
Additional information from the double-stranded DNA (dsDNA) Naldaviricetes phylogenetic analysis in Figure 5, including the best partition models chosen for each partition and the number of genes used for each species of the tree.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp7-v1.xlsx
-
Supplementary file 8
Biosample information regarding the 34 Hymenoptera species sequenced for this study.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp8-v1.xlsx
-
Supplementary file 9
Table representing the overlap between transposable elements and the clusters of homologous endogenous viral elements (EVEs).
The transposable elements were inferred using the RepeatModeler RepeatPeps database.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp9-v1.xlsx
-
Supplementary file 10
Information on the RNAseq datasets used in this study.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp10-v1.xlsx
-
Supplementary file 11
Details of the tblastn analysis for Platygaster orseoliae endogenous viral elements (PoEFV) and complementary information.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp11-v1.xlsx
-
Supplementary file 12
File including all the cluster phylogenies.
Leafs highlighted in green represent endogenous viral elements (EVEs) (scored from A to D), while leafs highlighted in red represent free-living viruses or loci annotated as putative free-living sequences (scored from D to X). The letter at the end of the taxon label represents the endogenization score assigned to the candidate. The assigned viral family of the free-living genes appears next to the pipe. The numbers right next to the ‘Event’ refers to the assigned Event number. If EVEs were found under selection (either by RNAseq or dN/dS analysis), the end of the leaf name will be ‘Selective_pressure_YES,’ while if not, the name will be ‘Selective_pressure_NO.’ Ultra-Fast Bootstrap values can be found next to the nodes of the phylogenies. For each phylogeny, the putative consensus protein name as well as the putative viral family is given at the top of the figure.
- https://cdn.elifesciences.org/articles/85993/elife-85993-supp12-v1.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/85993/elife-85993-mdarchecklist1-v1.pdf





