Modulation of protein-DNA binding reveals mechanisms of spatiotemporal gene control in early Drosophila embryos
Figures
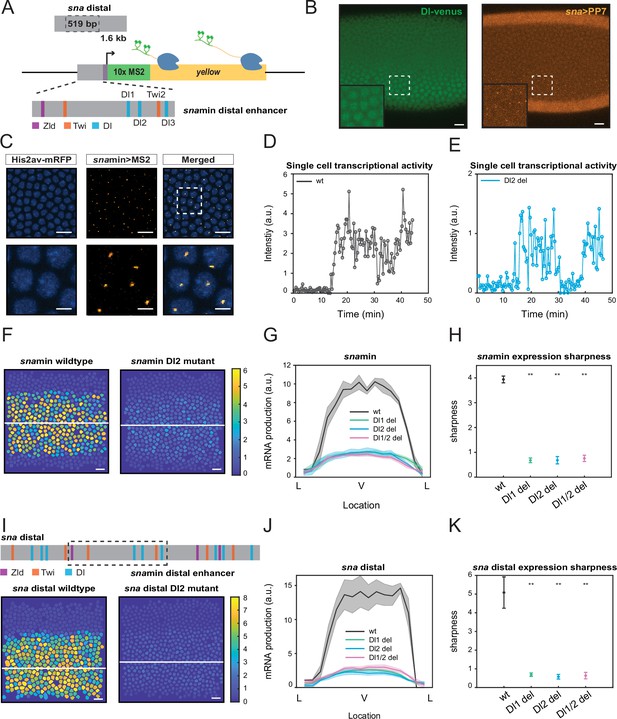
Single TF binding site mutation greatly diminishes mRNA production from the distal sna enhancer.
(A) Schematic of the reporter construct containing the minimal sna distal enhancer, sna core promoter, MS2 stem loops, and the yellow reporter gene. The minimal enhancer contains binding sites for TFs Zld, Twi, and Dl. (B) Embryo expressing maternally deposited Dl-Venus protein (green) and sna distal>PP7-yellow reporter gene (orange). sna is expressed in the region with high nuclear Dl. The inset shows the region within the rectangle. Scale bar = 10 μm. (C) Snapshots of an embryo expressing minimal sna distal>MS2-yellow. The nuclei are marked with Histone-RFP (blue) and the MS2-yellow reporter gene is observed with MCP-GFP (orange). Each nucleus has one distinct fluorescent punctum, indicating nascent transcription. Scale bar = 10 μm. The bottom row are magnifications of the embryo within the rectangle. Scale bar = 5 μm. (D–E) Single nucleus transcriptional trajectories for a wildtype (D) and Dl2 mutant embryo (E). (F) Heatmap showing that mRNA production is higher in a wildtype embryo compared to a Dl2 mutant embryo. The white line indicates the ventral midline. Scale bar = 12 μm. (G) Average mRNA production of all nuclei in wildtype embryos and Dl1, Dl2, and Dl1/2 mutant embryos across the sna expression domain (Lateral-Ventral-Lateral). (H) Plot indicating the wildtype steepness of snamin expression is significantly higher than the mutants. Steepness was determined by calculating the maximum derivative of the mRNA output curves. (I) Top: Schematic showing the additional TF binding sites present in the full sna distal enhancer. Bottom: Heatmap showing higher mRNA production of MS2-yellow in a wildtype embryo containing the full sna distal enhancer compared to the embryo containing the full enhancer with Dl2 site mutations. Scale bar = 12 μm. (J) Average mRNA production of all nuclei in wildtype embryos and Dl1, Dl2, and Dl1/2 mutant embryos containing the full sna distal enhancer across the expression domain. (K) Plot indicating the wildtype steepness of sna distal expression is significantly higher than the mutants. Shaded error bars in (G) and (J) indicate the standard error of the mean (SEM). Error bars in (H) and (K) indicate SEM. A total of 1524 nuclei from three replicate wildtype embryos, 1672 nuclei from four replicate Dl1 mutant embryos, 2091 nuclei from four replicate Dl2 mutant embryos, and 1788 nuclei from three replicate Dl1/2 mutant embryos were analyzed. ** denote p<0.001 from the student’s t-test.

Mutations affect mRNA production and expression sharpness, not width of expression domain.
(A) Average mRNA production of all nuclei in wildtype embryos and Dl3, Twi2, and Dl3Twi2 mutant embryos across the snamin expression domain. (B) Plot showing the width of the expression domain driven by snamin expression. There is no significant difference across the wildtype and mutant embryos. (C) Plot indicating the wildtype sharpness of snamin expression is significantly higher than the mutants. Shaded error bars in indicate standard error of the mean (SEM). Error bars in (B) and (C) indicate SEM. A total of 1524 nuclei from three replicate wildtype embryos, 1407 nuclei from three replicate Dl3 mutant embryos, 1557 nuclei from three replicate Twi2 mutant embryos, and 2058 nuclei from four replicate Dl3Twi2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the minimal sna distal enhancer. ** denote p<0.001 from the student’s t-test.
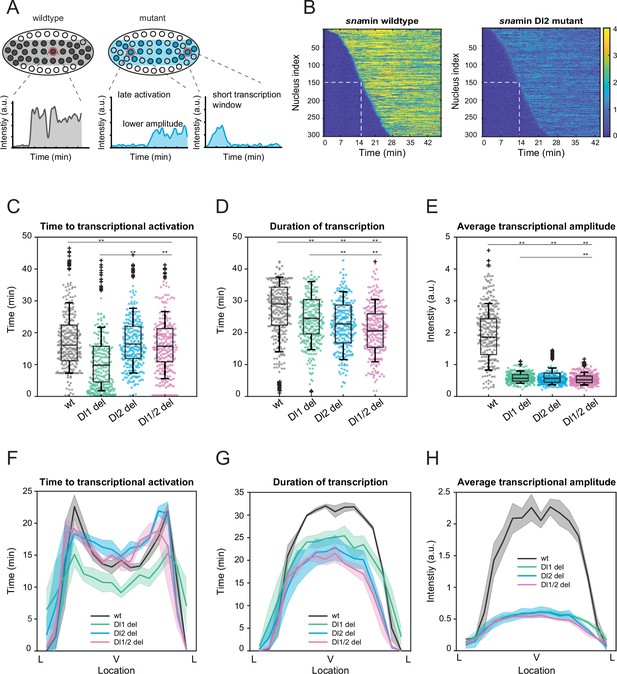
Mutations cause lower mRNA production, mainly due to lower transcriptional amplitude.
(A) Schematic comparing wildtype and mutants. The mutant embryo may have fewer nuclei transcribing (shaded) and at a lower intensity. Each nucleus in mutant embryos may have late activation or a shorter transcription window, all of which may contribute to the observed low mRNA production. (B) Heatmap of transcription activation times for representative wildtype and Dl2 mutant embryos. The time at which half of the nuclei are activated is indicated by the dotted white line and there is no significant difference. (C–E) Boxplots showing (C) the time to transcriptional activation (D) the duration of active transcription, and (E) the transcriptional amplitude for all actively transcribing nuclei. Decreased transcriptional amplitude contributes the most to the low mRNA production in mutants. (F–H) (F) Average time to transcriptional activation, (G) average duration of transcription for all actively transcribing nuclei, and (H) average transcriptional amplitude for all nuclei across the sna expression domain (Lateral – Ventral – Lateral). Nuclei in the middle of the expression domain are affected more, but there is no significant change in the expression width. Shaded error bars in (F–H) indicate SEM. 250 individual data points are overlaid on the respective boxplots. A total of 1124 nuclei from three replicate wildtype embryos, 1011 nuclei from four replicate Dl1 mutant embryos, 1123 nuclei from four replicate Dl2 mutant embryos, and 943 nuclei from three replicate Dl1/2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the minimal sna distal enhancer. ** denote p<0.001 from the student’s t-test.

Mutations cause lower mRNA production, mainly due to lower transcriptional amplitude.
(A–C) Boxplots showing (A) the time to transcriptional activation, (B) the duration of active transcription, and (C) the transcriptional amplitude for all actively transcribing nuclei. Decreased transcriptional amplitude contributes the most to the low mRNA production in mutants. (D–F) (D) Average time to transcription activation, (E) average duration of transcription for all actively transcribing nuclei, and (F) average transcriptional intensity for all nuclei across the sna expression domain (Lateral – Ventral – Lateral). Nuclei in the middle of the expression domain are affected more, but there is no significant change in the expression width. Shaded error bars in (D–F) indicate SEM. 250 individual data points are overlaid on the respective boxplots. A total of 1124 nuclei from three replicate wildtype embryos, 828 nuclei from three replicate Dl3 mutant embryos, 754 nuclei from three replicate Twi2 mutant embryos, and 983 nuclei from four replicate Dl3Twi2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the minimal sna distal enhancer. ** denote p<0.001 from the student’s t-test comparing wildtype and mutant embryos.
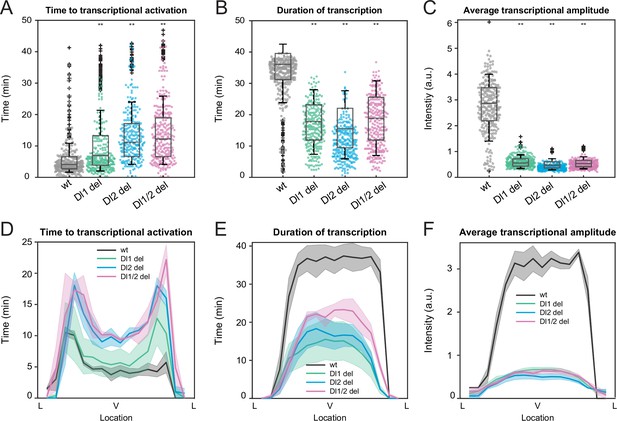
Mutations in the full sna distal enhancer decrease duration of transcription in addition to lowering transcriptional amplitude.
(A–C) Boxplots showing (A) the time to transcriptional activation, (B) the duration of active transcription, and (C) the transcriptional amplitude for all actively transcribing nuclei. Decreased transcriptional amplitude contributes the most to the low mRNA production in mutants. (D–F) (D) Average time to transcriptional activation, (E) average duration of transcription for all actively transcribing nuclei, and (F) average transcriptional intensity for all nuclei across the sna expression domain (Lateral – Ventral – Lateral). Nuclei in the middle of the expression domain are affected more, but there is no significant change in the expression width. Shaded error bars in (D–F) indicate SEM. A total of 730 nuclei from two replicate wildtype embryos, 1567 nuclei from four replicate Dl1 mutant embryos, 1030 nuclei from three replicate Dl2 mutant embryos, and 981 nuclei from three replicate Dl1/2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the full sna distal enhancer. ** denote p<0.001 from the student’s t-test comparing wildtype and mutant embryos.
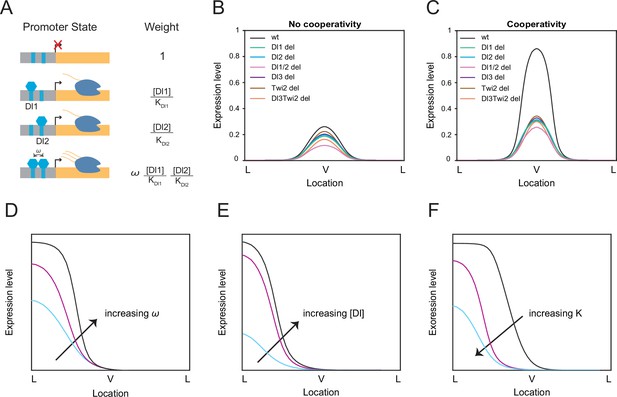
Thermodynamic equilibrium binding model reveals synergy among TF binding sites.
(A) Promoter states and statistical weights for each microstate. A bound activator will yield transcription. Cooperativity term ω is included when more than one TF is bound, which will result in higher mRNA production. Dissociation constants are given by Ki, which is correlated with the binding affinity of each site. (B–C) mRNA production curves generated by assuming (B) no cooperativity and (C) cooperativity among TF binding sites. Modeling results support experimental data with the cooperativity term included. (D–F) Summary models noting expression level changes as cooperativity increases (D), as Dl concentration is increased in low binding affinity enhancers (E), and as binding affinity is increased (F).
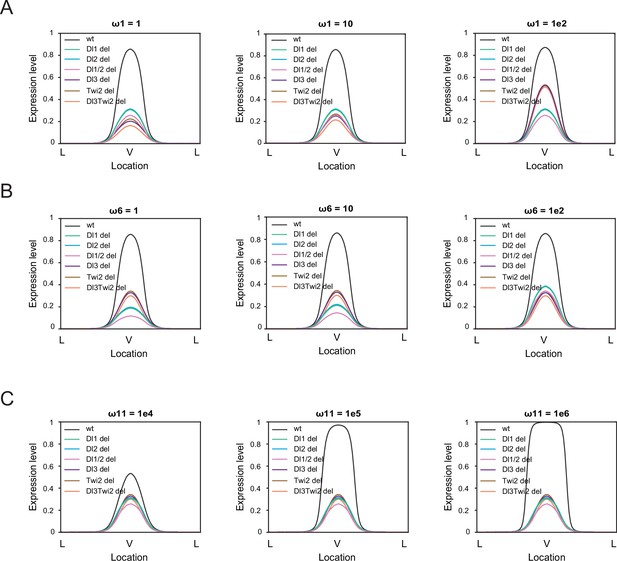
Sensitivity analysis on thermodynamic model highlights the importance of cooperativity among all TFs.
(A–B) Sensitivity analysis for ω1 (A) and ω6 (B) for values of 1 (left), 10 (center), and 1e2 (right). The model is robust against changes in these parameters. (C) Sensitivity analysis for ω11 for values of 1e4 (left), 1e5 (center), and 1e6 (right) indicates the model is more sensitive to changes in cooperativity among all 4 TFs.
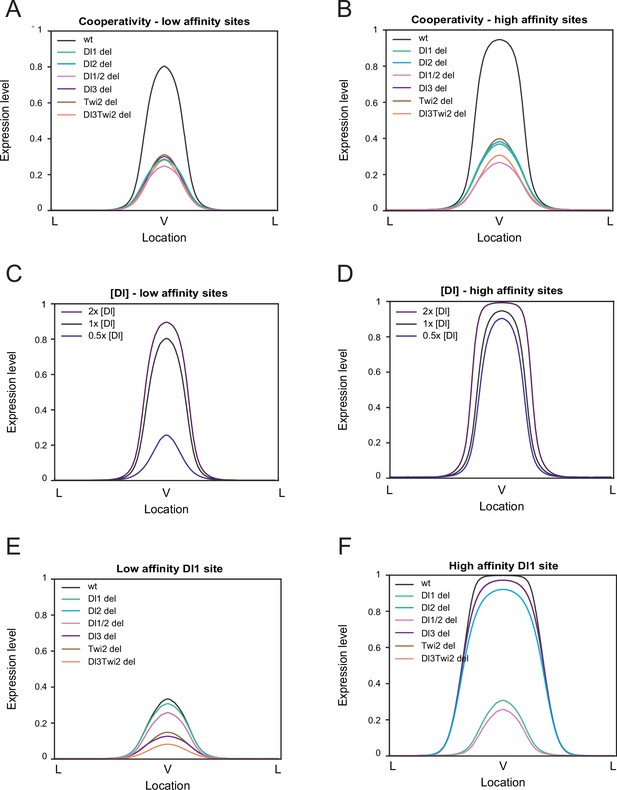
Modulation of binding affinity and Dl concentration.
(A–B) Model results of low-affinity sites (A) and high affinity sites (B) with cooperativity. Higher cooperativity values are needed for the low-affinity model (see Table 2). (C–D) Modulation of Dl concentration with low-affinity sites (C) and with high-affinity sites (D). Lowering Dl concentration causes a drastic reduction in expression levels in enhancers with low-affinity sites. (E–F) Altering affinity of a single site, Dl1, to low affinity (E) and high affinity (F). Decreasing affinity severely reduces expression while increasing affinity results in wider and ectopic expression.
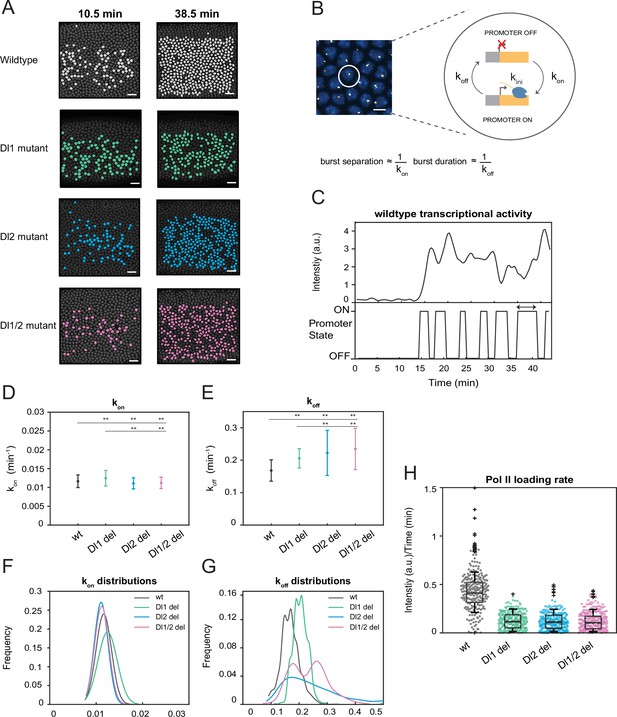
A two-state model reveals differences in koff rates and burst duration.
(A) Actively transcribing nuclei are false-colored for early and late NC14. Mutant embryos show more sporadic transcriptional activity in a given frame. Scale bar = 15 μm. (B) Schematic depicting a single nucleus that can be in an OFF or ON state and switches between the two with rates kon and koff. The rates of kon and koff can be correlated to burst separation and burst duration, respectively. Scale bar = 5 μm. (C) Representative transcriptional trajectory of wildtype with the inferred promoter states derived from the Hidden Markov model (HMM). Arrow indicates burst duration. (D–E) Plots showing the rates of (D) kon and (E) koff. kon is not significantly affected, whereas koff rates are higher in the mutant embryos. (F–G) Probability distributions of (F) kon rates and (G) koff rates for wildtype, Dl1, Dl2, and Dl1/2 mutants. The distribution of kon rates follow a tight normal distribution while the koff distributions vary widely. (H) Boxplot showing the Pol II loading rates of all actively transcribing nuclei. The wildtype embryos have a significantly higher rate of Pol II loading than the mutant embryos. Error bars in (D–E) indicate standard deviation (SD). A total of 1124 nuclei from three replicate wildtype embryos, 1011 nuclei from four replicate Dl1 mutant embryos, 1123 nuclei from four replicate Dl2 mutant embryos, and 943 nuclei from three replicate Dl1/2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the minimal sna distal enhancer. ** denote p<0.001 from the student’s t-test.
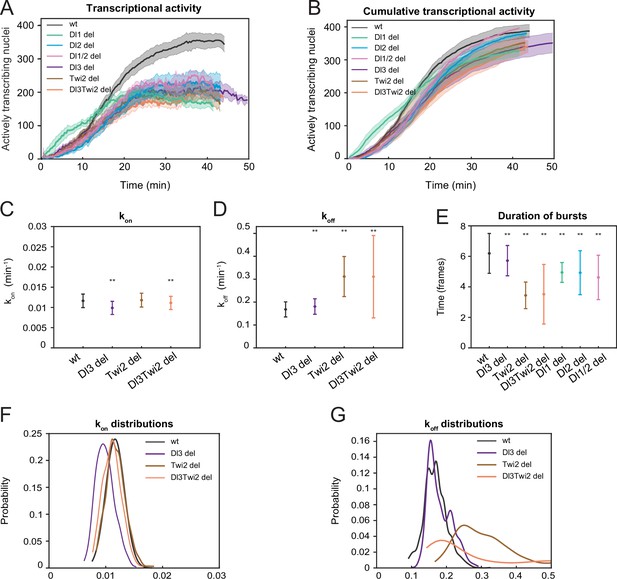
A two-state model reveals differences in koff rates and burst duration.
(A) Plot showing the number of actively transcribing nuclei at every timepoint for wildtype and all mutant embryos. In the wildtype, the number of nuclei steadily increases, while in the mutant, the number of active nuclei at any given timepoint is much lower. (B) Plot showing the cumulative number of actively transcribing nuclei. The number of nuclei is comparable across the wildtype and mutant embryos. (C–E) Plots showing the rates of (C) kon, (D) koff, and (E) burst duration for Dl3, Twi2, and Dl3Twi2 mutants. kon is not significantly affected, whereas koff rates are higher in the mutant embryos. Burst duration is also reduced in all the mutant embryos. (F–G) Probability distributions of (F) kon rates and (G) koff rates for wildtype, Dl3, Twi2, and Dl3Twi2 mutants. The distribution of kon rates follow a tight normal distribution while the koff distributions vary widely. Shaded error bars in (A–B) indicate SEM. Error bars in (C–E) indicate standard deviation (SD). A total of 1124 nuclei from three replicate wildtype embryos, 828 nuclei from three replicate Dl3 mutant embryos, 754 nuclei from three replicate Twi2 mutant embryos, and 983 nuclei from four replicate Dl3Twi2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the minimal sna distal enhancer. ** denote p<0.001 from the student’s t-test comparing wildtype and mutant embryos.

Mutations in the full sna distal enhancer increase koff rates leading to reduced burst duration.
(A) Plot showing the number of actively transcribing nuclei at every timepoint for wildtype and all mutant embryos. In the wildtype, the number of nuclei steadily increases, while in the mutant, the number of active nuclei is much more stochastic at any given timepoint. (B–C) Probability distributions of (B) kon rates and (C) koff rates for wildtype, Dl1, Dl2, and Dl1/2 mutants. The distribution of kon rates follow a tight normal distribution while the koff distributions vary widely. (D–F) Plots showing the rates of (D) kon, (E) koff, and (F) burst duration for Dl1, Dl2, and Dl1/2 mutants. kon is not significantly affected, whereas koff rates are higher in the mutant embryos. Burst duration is also reduced in all the mutant embryos. Shaded error bars in (A) indicate SEM. Error bars in (D–F) indicate SD. A total of 730 nuclei from two replicate wildtype embryos, 1567 nuclei from four replicate Dl1 mutant embryos, 1030 nuclei from three replicate Dl2 mutant embryos, and 981 nuclei from three replicate Dl1/2 mutant embryos were analyzed. Transcription of the MS2-yellow reporter gene is driven by the full sna distal enhancer. ** denote p<0.001 from the student’s t-test comparing wildtype and mutant embryos.
Videos
Live imaging of wildtype embryo expressing snamin > MS2-yellow during NC14.
MS2 signal is shown in yellow. Nuclei are marked with His2Av-mRFP. Histogram was adjusted for visualization purposes. Scale bar = 10 μm.
Live imaging of Dl2 mutant embryo expressing snamin > MS2-yellow during NC14.
MS2 signal is shown in yellow. Nuclei are marked with His2Av-mRFP. Scale bar = 10 μm.
False coloring of actively transcribing nuclei of a wildtype snamin> MS2-yellow embryo.
Scale bar = 10 μm.
False coloring of actively transcribing nuclei of a Dl2 mutant snamin> MS2-yellow embryo.
Scale bar = 10 μm.
Tables
Table of TF motifs and mutated sequences.
Table showing the sequences of the TF motifs, p-value, mutated sequences, and Patser score.
| Transcription factor | p-value | Patser score | Wildtype | Mutation |
|---|---|---|---|---|
| Dl1 | 1.43e-4 | 22.8 | AGGGATTTCCT | AGGGATCGCCT |
| Dl2 | 2.71e-05 | 19.8 | GGCGTTTTCCCA | GGCGATTGACCA |
| Dl3 | 8.52e-05 | 17.8 | TGGGAAATCGG | TGTTAAATCGG |
| Twi2 | 4.08e-05 | 7.8 | GTCCATGTGTTG | GTCCATGAATTG |
Table of cooperativity values.
Table showing the cooperativity values for the optimized model and the cooperativity values when all binding sites are weakened or strengthened. Cooperativity values are much higher when the strength of the binding sites are weakened.
| Cooperativity term | Transcription factor | Optimized value | Weak binding value | Strong binding value |
|---|---|---|---|---|
| ω1 | Dl1, Dl2 | 28.00 | 69.90 | 2.03 |
| ω2 | Dl2, Twi2 | 1.00 | 1.00 | 1.00 |
| ω3 | Dl1, Twi2 | 1.00 | 1.00 | 1.00 |
| ω4 | Dl1, Dl3 | 1.00 | 1.00 | 1.00 |
| ω5 | Dl2, Dl3 | 1.01 | 1.00 | 1.00 |
| ω6 | Dl3, Twi2 | 55.30 | 139.00 | 6.79 |
| ω7 | Dl1, Dl2, Twi2 | 1.07 | 1.05 | 1.03 |
| ω8 | Dl1, Dl2, Dl3 | 1.03 | 1.02 | 1.01 |
| ω9 | Dl2, Twi2, Dl3 | 1.13 | 61.10 | 1.03 |
| ω10 | Dl1, Twi2, Dl3 | 1.03 | 1.03 | 1.02 |
| ω11 | Dl1, Dl2, Twi2, Dl3 | 1.59e5 | 5.00e5 | 3.10e4 |
Table of promoter occupancies with and without cooperativity.
Table showing the promoter occupancies in the cases of cooperativity and no cooperativity for the three cooperativity terms that were greater than 1 in the optimized model. Cooperativity increases promoter occupancy by 90%.
| Promoter state | Promoter occupancy with cooperativity | Promoter occupancy with no cooperativity |
|---|---|---|
| ω1 | 2.4378 | 0.0870 |
| ω6 | 2.1913 | 0.0397 |
| ω11 | 54.8073 | 3.45e-04 |





