Recapitulation of pathophysiological features of AD in SARS-CoV-2-infected subjects
Figures
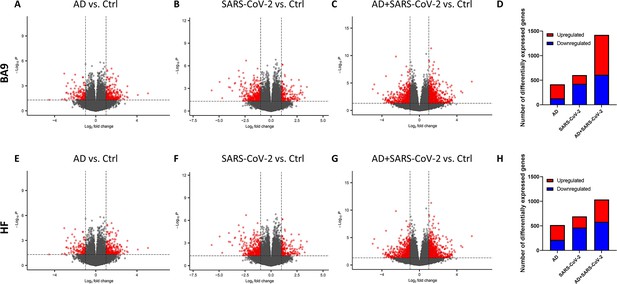
Gene expression in cortical Brodmann area 9 (BA9) and hippocampal formation (HF).
(A–C) Volcano plot distribution of gene transcripts of the cortical BA9 region in Alzheimer’s disease (AD) cases (A), SARS-CoV-2 cases (B), and SARS-CoV-2-infected AD cases (C) compared to neurological control cases. (E–G) Volcano plot distribution of gene transcripts of the cortical HF region in AD cases (E), SARS-CoV-2 cases (F), and SARS-CoV-2-infected AD cases (G) compared to neurological control cases. Volcano plots were generated from 39,901 genes (A, B, E, F), 38,021 genes (C), and 35,527 genes (G). Transcripts with nominal p<0.01 and an absolute log2 fold-change (log2 FC) > 1 are indicated in red. (D, H) Bar graphs, in which the number of upregulated and downregulated genes (with p<0.01 and absolute value of log2 FC > 1), is indicated in red and blue, respectively.
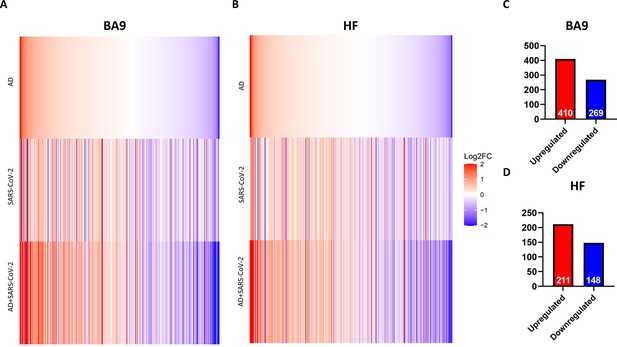
Similarity of gene expression in cortical Brodmann area 9 (BA9) and hippocampal formation (HF) in SARS-CoV-2 and SARS-CoV-2-infected Alzheimer’s disease (AD) relative to AD individuals based on log2 fold-change (log 2FC).
(A, B) Genes known to be differentially expressed due to either SARS-CoV-2 infection or AD were examined with respect to their gene expression changes, with the given gene set being selected. The log2 FC of the genes for three individual differential comparisons (AD versus control, SARS-CoV-2 versus control, and SARS-CoV-2-infected AD versus control) are shown for cortical BA9 (A) and HF (B), respectively. (C, D) Using Ingenuity Pathway Analysis (IPA) as a filtering system for genes within the database, we compared the similarity in expression of genes present in all datasets (SARS-CoV-2, AD and SARS-CoV-2-infected AD cases) in both the cortical BA9 (C) and HF (D).
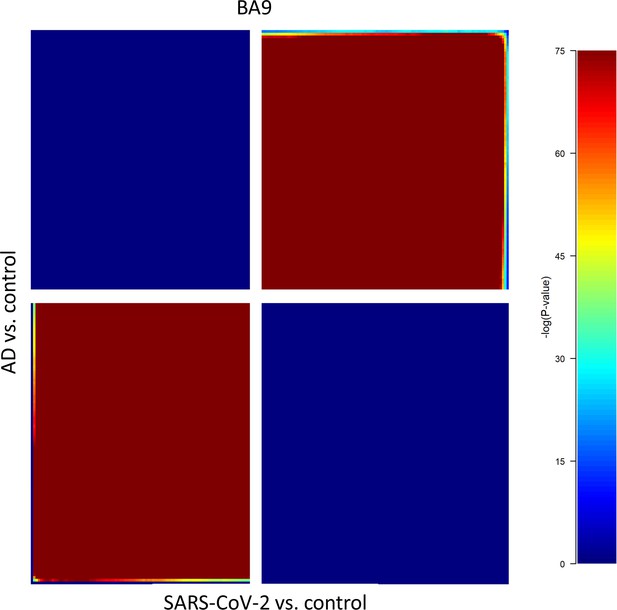
Rank-rank hypergeometric overlap (RRHO) analysis of SARS-CoV-2/control and Alzheimer’s disease (AD)/control, each containing (39,902 differentially expressed genes [DEGs]) revealed that gene regulation between the AD/control and SARS-CoV-2/control groups showed a positive correlation in the cortical Brodmann area 9 (BA9) region.
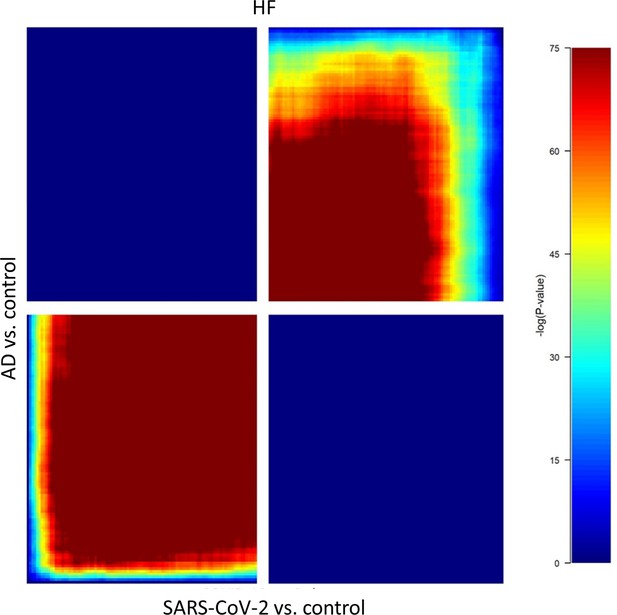
Rank-rank hypergeometric overlap (RRHO) analysis of SARS-CoV-2/control and Alzheimer’s disease (AD)/control, each containing (39,902 differentially expressed genes [DEGs]) revealed that gene regulation between the AD/control and SARS-CoV-2/control groups showed a positive correlation in the HP region.
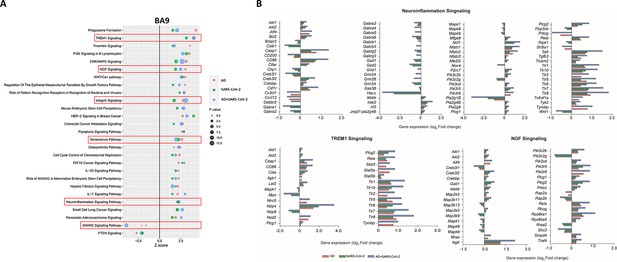
Changes in signaling pathways within the cortical Brodmann area 9 (BA9).
(A) The similarities of canonical pathways reveal the top regulated canonical pathways within the cortical BA9 in Alzheimer’s disease (AD), SARS-CoV-2, and SARS-CoV-2-infected AD cases in reference to the control. Activation score (Z-score) is shown on the X-axis, while the pathways are indicated on the Y-axis. The color of the points indicates the Ingenuity Pathway Analysis (IPA) comparison, while the size of the point represents the -log10 p-value of the IPA comparison, with the larger points indicating the lowest p-values. (B) The predicted gene regulation of the Neuroinflammation, TREM1, and nerve growth factor (NGF) pathways indicates that AD (red bar), SARS-CoV-2 (green bar), and SARS-CoV-2-infected AD groups (blue bar) have similar expression in key inflammatory and neuronal pathways, with log2 fold-change (log2 FC) shown on the X-axis.
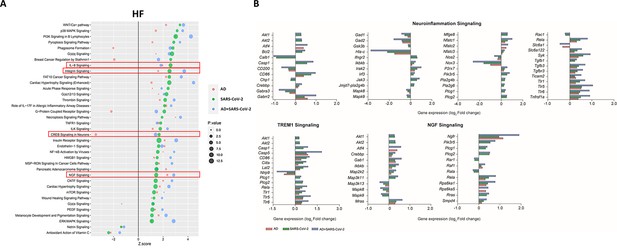
Changes in signaling pathways within the hippocampal formation (HF).
(A) The similarities of canonical pathways reveal the top regulated canonical pathways were compared within the HF in Alzheimer’s disease (AD), SARS-CoV-2, and SARS-CoV-2-infected AD cases in reference to the control. The X-axis represents the activation score (Z-score), while the Y-axis indicates the pathways. The color of the points reflects the Ingenuity Pathway Analysis (IPA) comparison, while the size of each point corresponds to the -log10 p-value of the IPA comparison, with larger points indicating lower p-values. (B) The predicted gene regulation of the Neuroinflammation, TREM1, and nerve growth factor (NGF) pathways indicates that AD (red bar), SARS-CoV-2 (green bar), and SARS-CoV-2-infected AD groups (blue bar) have similar expression in key inflammatory and neuronal pathways. The X-axis represents log2 fold-change (log2 FC).
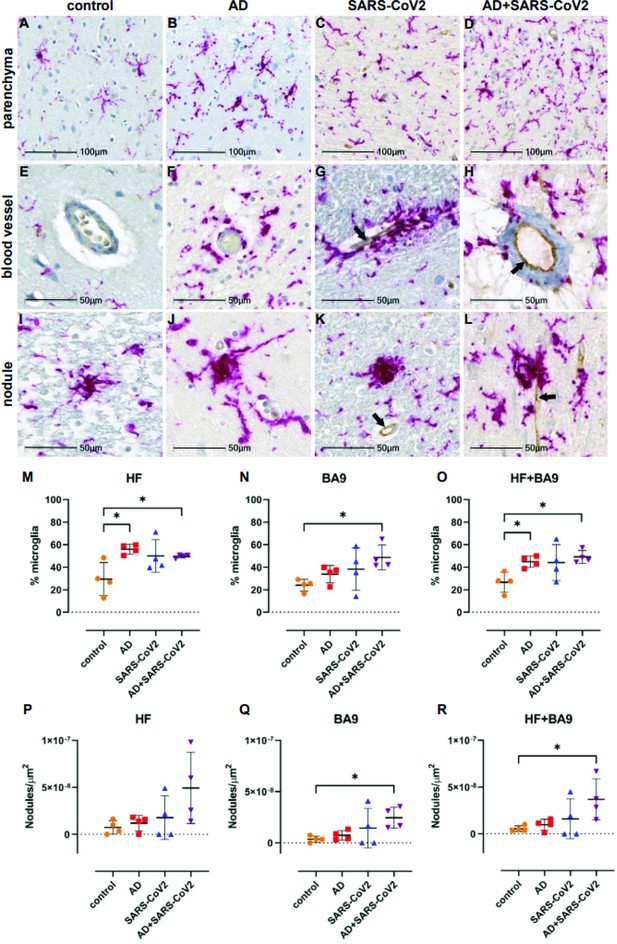
Microgliosis and nodular lesions in neurological controls, SARS-CoV-2, Alzheimer’s disease (AD), and SARS-CoV-2-infected AD individuals.
The level of microglial activation was assessed by immunohistochemical staining using anti-Iba-1 antibody with Vector Red. The presence of SARS-CoV-2 was assessed using an antibody against the virus nucleocapsid and visualized with DAB. (B–D) Parenchymal microglia are more frequent and highly activated in the context of infection with and without AD, as shown by thickened processes, enlarged soma, and loss of individual cellular domain, compared to age-matched controls (A, E). When present, SARS-CoV-2 localizes to the blood vessel endothelium (black arrows; G, H, K, L). (F–H) Microglia appear to gather around blood vessels in disease but do not form cuffs. (I–L) Nodular lesions are seen in most cases assessed, regardless of disease status; however, they appear larger and more frequent in the context of disease (J–L), compared to controls (I). (M–O) A multiplex algorithm was used to count all cells, using DAPI+ nuclei, with HALO and calculate percent frequency of Iba-1+ microglia. Each point on the graphs shown represents the average finding per total brain area for each subject. (M) There was upregulation of Iba-1 in the hippocampus compared to the cortical Brodmann area 9 (BA9) region for most cases, though there is a significantly higher ratio of microglia relative to other cells when comparing the AD and SARS-CoV-2-infected AD cases to the control group shown. (N) This trend seems to hold true for cortical BA9 region as well, where the only groups with a significant difference in microglia ratios are the control and SARS-CoV-2-infected AD cases shown. (O) When all cases have both regions averaged, the level of microgliosis is shown to be higher in the AD and SARS-CoV-2-infected AD cases compared to control. (P–R) Graphs show the normalized counts of microglial nodule frequency. (P) A higher frequency of nodules is seen in the HF; however, the difference between groups did not reach statistical significance. (Q) In contrast, fewer nodules are seen in cortical BA9, overall. A statistically significant higher number of lesions are seen in SARS-CoV-2-infected AD cases compared to control cortical BA9, suggesting greater inflammation in the cortical BA9 of patients with both AD and SARS-CoV-2 infection. (R) Significance was maintained between the control and SARS-CoV-2-infected AD groups when the average was taken for both brain regions per case. Statistics were performed with a two-tailed Mann–Whitney U-test. *p<0.05. Data are expressed as mean ± SEM.
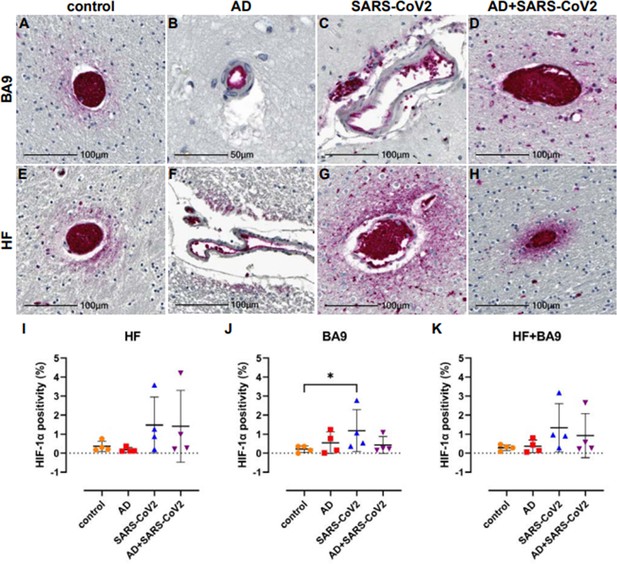
Hypoxia-inducible factor-1 alpha subunit (HIF-1α) in neurological controls, SARS-CoV-2, Alzheimer’s disease (AD), and SARS-CoV-2-infected individuals.
(A–H) Representative images demonstrate HIF-1α immunopositivity around the vasculature that extends into the parenchyma in brain of patients with SARS-CoV-2 infection (C, D, G, H), regardless of AD status. Comparatively, this is seen less frequently and does not extend significantly into the parenchyma in brain of age-matched controls (A, E). In AD only, positivity was most often observed in epithelium with no or minimal extension into the brain parenchyma (B, F). (I–K) Graphs show the total average percentage of tissue positive for HIF-1α for each individual subject, as defined using a HALO area algorithm for detection of Vector Red intensity over the whole section. Although statistical significance between groups was not reached in the hippocampal formation (HF) (I) or averaged group comparisons (K), increased HIF-1α expression is seen in the SARS-CoV-2-infected patients, with and without AD, compared to age-matched controls. An increase in area positivity is seen in all groups in the cortical Brodmann area 9 (BA9) region (J), compared to nonaffected controls; however, statistical significance is only seen with the SARS-CoV-2 group. Statistics were performed with a two-tailed Mann–Whitney U-test. *p<0.05. Data are expressed as mean ± SEM.
Additional files
-
Supplementary file 1
Table 1. Demographics of postmortem cases.
Numbers presented are the average per group and standard deviation PMI is postmortem interval (n = 4); OtD is onset to death. ABC is a multipoint measurement where A is a measure of amyloid β deposition, B is a measure of neurofibrillary degeneration based on the Braak score, and C is scored based on neuritic plaques outlined by the Consortium to Establish a Registry for Alzheimer’s Disease diagnosis (CERAD).
- https://cdn.elifesciences.org/articles/86333/elife-86333-supp1-v2.xlsx
-
Supplementary file 2
Table 2. Blood chemistry and symptoms.
Symptoms are presented as a percentage of the group (n = 4); chemistry numbers are presented as the average per group and standard deviation, based on maximum values collected for single values and the minimum and maximum range collected in cells with two values. Bolded values are implicated as indicators of severe SARS-CoV-2 outcomes; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate transferase; CRP, c-reactive protein; ESR, erythrocyte sedimentation; INR, international normalized ratio; LDH, lactate dehydrogenase; WBC, white blood count; BUN, blood urea nitrogen.
- https://cdn.elifesciences.org/articles/86333/elife-86333-supp2-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86333/elife-86333-mdarchecklist1-v2.pdf
-
Source data 1
Based on the DESeq, and filtered using IPA, genes that were expressed in all SARS-CoV-2 and AD in cortical BA9 datasets with an absolute expression of 0.0001 or greater and p<0.05 are presented.
- https://cdn.elifesciences.org/articles/86333/elife-86333-data1-v2.xlsx
-
Source data 2
Based on the DESeq, and filtered using IPA, genes that were expressed in all SARS-CoV-2 and AD in HF datasets with an absolute expression of 0.0001 or greater and p<0.05 are presented.
- https://cdn.elifesciences.org/articles/86333/elife-86333-data2-v2.xlsx





