PerTurboID, a targeted in situ method reveals the impact of kinase deletion on its local protein environment in the cytoadhesion complex of malaria-causing parasites
Figures
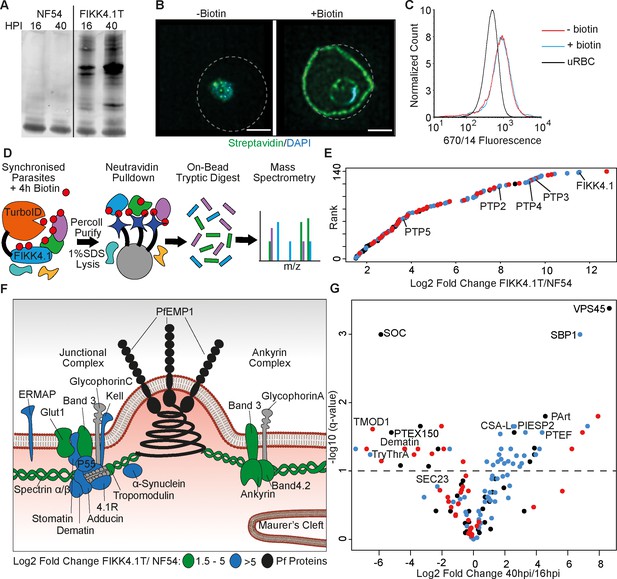
Proximity-dependent protein labeling of the FIKK4.1 subcellular niche.
(A) Western blot of parental NF54 and the FIKK4.1-TurboID (FIKK4.1T) fusion line probed with streptavidin conjugated to a fluorophore. Parasites were incubated with biotin either between 16–20 hr post-infection (hpi) or 40–44 hpi. (B) Structured illumination microscopy of FIKK4.1T fusion line incubated with or without biotin, and probed with streptavidin-fluorophore (green), and DAPI (blue). Dotted lines represent the approximate position of the RBC membrane according to brightfield images (see Figure 1—figure supplement 3). Scale bar: 2 µm. (C) Flow cytometric analysis of FIKK4.1T fusion lines using anti-Var2CSA antibodies. (D) Schematic of the classical TurboID protocol. TurboID bait fusion parasites are incubated with biotin (red), purified, then lysed. Biotinylated proteins are enriched, digested, and peptides quantified using mass spectrometry. (E) Ranked plot of all proteins quantified above NF54 background (x-axis – log2 fold change FIKK4.1T/NF54). Red, human proteins; blue, exported parasite proteins; black, non-exported parasite proteins. (F) Schematic of a knob structure at the infected red blood cell (iRBC) cytoskeleton. Human proteins with log2 fold change FIKK4.1T/ NF54 from 1.5 to 5 are highlighted in green, and the most abundant proteins with values >5 are in blue. Parasite proteins are in black, and other RBC proteins are in gray (G) Volcano plot depicting the log2 fold change in abundance between early (16–20 hpi) and late (40–44 hpi) biotin pulses with FIKK4.1T parasites. Red, human proteins; blue, exported P. falciparum proteins; black, non-exported P. falciparum proteins. Selected proteins are labeled.
-
Figure 1—source data 1
Unedited blot for Figure 1A.
- https://cdn.elifesciences.org/articles/86367/elife-86367-fig1-data1-v2.zip
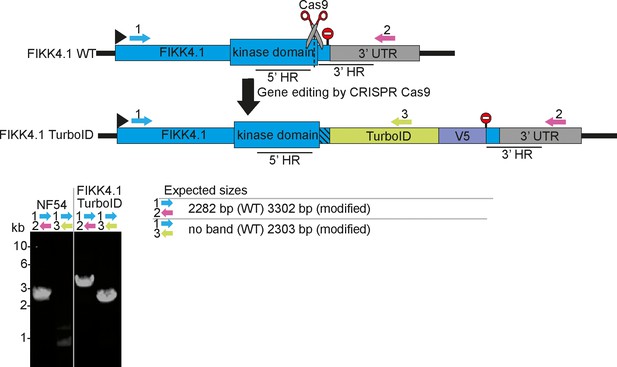
PCRs confirming correct genome modification of the FIKK4.1 locus.
Primers are represented by numbered arrows on the schematics, stop codons by stop signs. Homology regions used to edit the genome are denoted by 5’ and 3’ HR, and Cas9 guide sites are denoted by scissors. Primer combinations, parasite lines, and whether rapamycin treatment was applied are denoted above each PCR blot, and the expected sizes of PCR bands are shown on the bottom right.
-
Figure 1—figure supplement 1—source data 1
Original data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/86367/elife-86367-fig1-figsupp1-data1-v2.zip
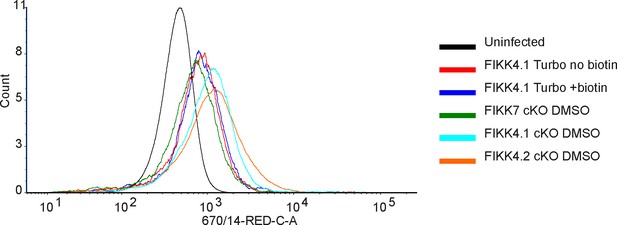
Verification of Var2CSA surface translocation in FIKK4.1-TurboID transgenic lines.
Flow cytometric analysis of FIKK4.1-TurboID, FIKK4.1 cKO, FIKK4.2 cKO, and FIKK7.1 cKO fusion lines using anti-Var2CSA antibodies. FIKK4.1-TurboID+ biotin was treated with biotin for 4 hr. This confirms that the addition of the TurboID tag to FIKK4.1 and the addition of biotin does not substantially impair Var2CSA surface translocation relative to other parasite lines.
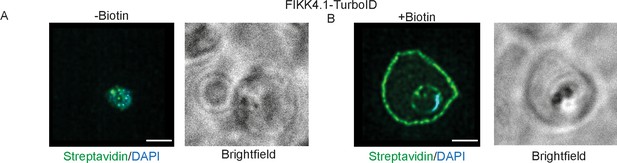
Structured illumination microscopy of FIKK4.1-TurboID lines with brightfield images included.
Related to images (A, B) from Figure 1B.
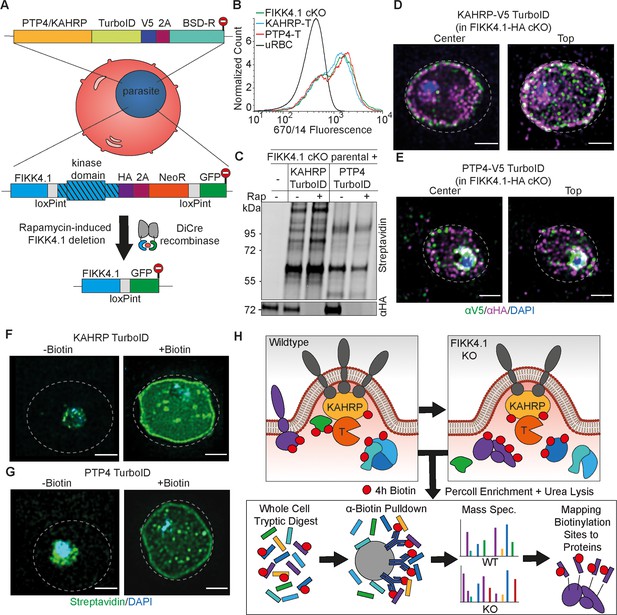
Design and validation of KAHRP and PTP4-TurboID tagging in a FIKK4.1 cKO line for PerTurboID.
(A) Schematic of the generation of double transgenic parasite lines containing a loxP-flanked FIKK4.1 kinase domain for conditional KO and KAHRP/PTP4-TurboID fusions. (B) Surface translocation of Var2CSA in KAHRP/PTP4-TurboID and parental lines analyzed by flow cytometry using anti-Var2CSA antibodies. (C) Western blot of FIKK4.1 cKO parental line and KAHRP/PTP4 TurboID-fusion parasites, with full-length FIKK4.1 (-RAP) or the kinase domain deleted (+RAP). The blot was probed with streptavidin-fluorophore or anti-HA antibodies targeting FIKK4.1-HA, confirming excision and biotinylation. (D, E) Structured illumination microscopy of KAHRP/PTP4:TurboID fusion lines probed with anti-HA (FIKK4.1, magenta), anti-V5 (PTP4 or KAHRP, green), and DAPI, blue. Views from the top and center of the cell are shown. (F, G) TurboID transgenic parasites incubated with or without biotin and probed with streptavidin-fluorophore (green) and DAPI (blue). Dotted lines represent the approximate position of the RBC membrane according to bright field images (see Figure 2—figure supplement 5). Scale bar: 2 µm. (H) Schematic depicting the PerTurboID protocol. FIKK4.1 cKO induces hypothetical changes to proteins in the vicinity of KAHRP/PTP4, leading to differences in the accessibility of lysines to biotinylation. Parasites are purified, lysed, then digested. Biotinylated peptides are enriched, then eluted and quantified by mass spectrometry. Biotinylated peptides are mapped onto the protein, indicating protein domains accessible to the TurboID biotin ligase. T, TurboID tag.
-
Figure 2—source data 1
Unedited blot for Figure 2C.
- https://cdn.elifesciences.org/articles/86367/elife-86367-fig2-data1-v2.zip
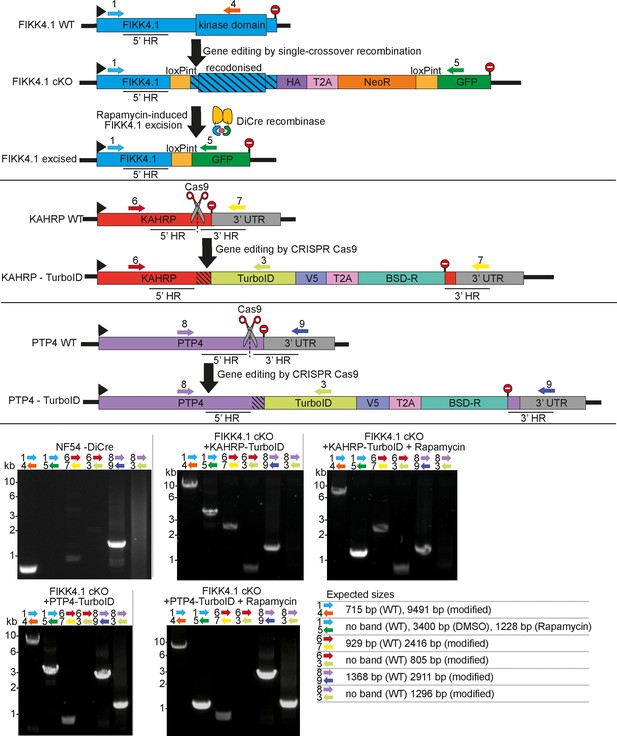
PCRs confirming correct genome modification of the PTP4 and KAHRP locus.
Primers are represented by numbered arrows on the schematics, stop codons by stop signs. Homology regions used to edit the genome are denoted by 5’ and 3’ HR, and Cas9 guide sites are denoted by scissors. Primer combinations, parasite lines, and whether rapamycin treatment was applied are denoted above each PCR blot, and the expected sizes of PCR bands are shown on the bottom right.
-
Figure 2—figure supplement 1—source data 1
Unedited DNA gels for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/86367/elife-86367-fig2-figsupp1-data1-v2.zip
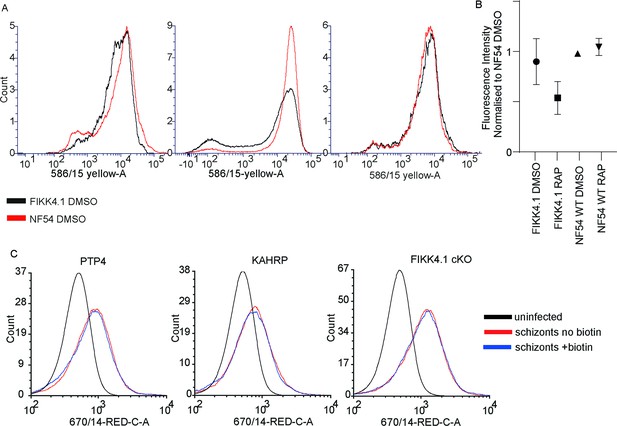
Verification of Var2CSA surface translocation in KAHRP and PTP4-TurboID transgenic lines.
(A) Flow cytometric verification that FIKK4.1 cKO displays similar levels of Var2CSA on the infected red blood cell (iRBC) surface compared to NF54 wildtype. Three replicates are shown, the median fluorescence intensity of which is shown in (B), normalized to the NF54 DMSO line. (C) Flow cytometric analysis of the effect of a 4 hr biotin treatment on Var2CSA surface translocation in the KAHRP-TurboID and PTP4-TurboID line, as well as the parental line FIKK4.1 cKO as a control.
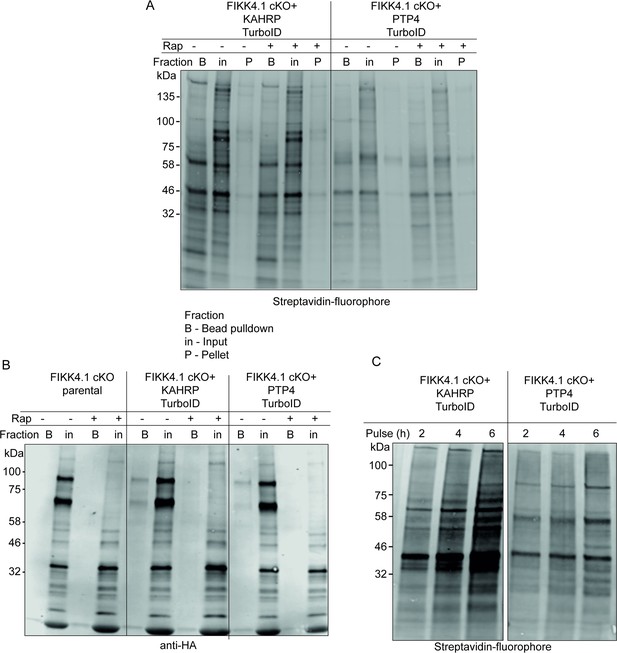
Western blot analysis of KAHRP and PTP4-TurboID fusion lines.
(A, B) Pulldown of biotinylated proteins by neutravidin-coated beads. Blots were probed with (A) streptavidin-fluorophore or (B) anti-HA targeting FIKK4.1-HA. FIKK4.1 was pulled down only in KAHRP/PTP4-TurboID untreated samples and not the parental line, indicating it is biotinylated by TurboID and successfully excised upon treatment with rapamycin. A double band is observed for FIKK4.1, likely representing a FIKK4.1-NeoR population due to incomplete T2A cleavage of exported proteins. (C) Increasing duration of biotin pulse on biotinylation of proteins in KAHRP/PTP4-TurboID lines. The blot was probed with streptavidin-fluorophore.
-
Figure 2—figure supplement 3—source data 1
Unedited blots for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/86367/elife-86367-fig2-figsupp3-data1-v2.zip
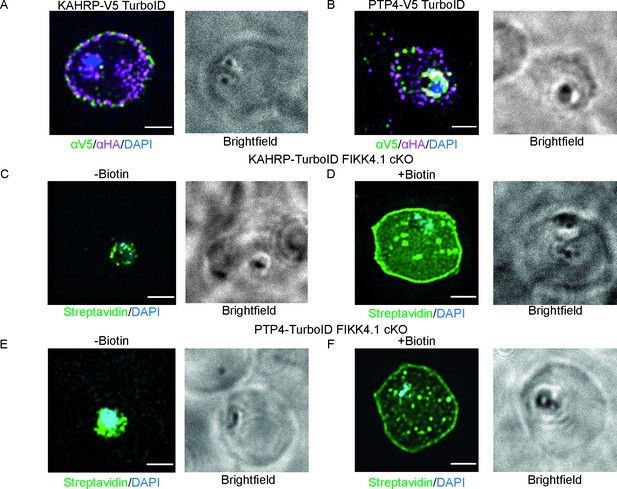
Structured illumination microscopy of KAHRP and PTP4-TurboID lines with brightfield images included.
Images (A) and (B) related to Figure 2C and D; (C) and (D) related to Figure 2F; and (E); (F) related to Figure 2G, Scale bar: 2 µm.
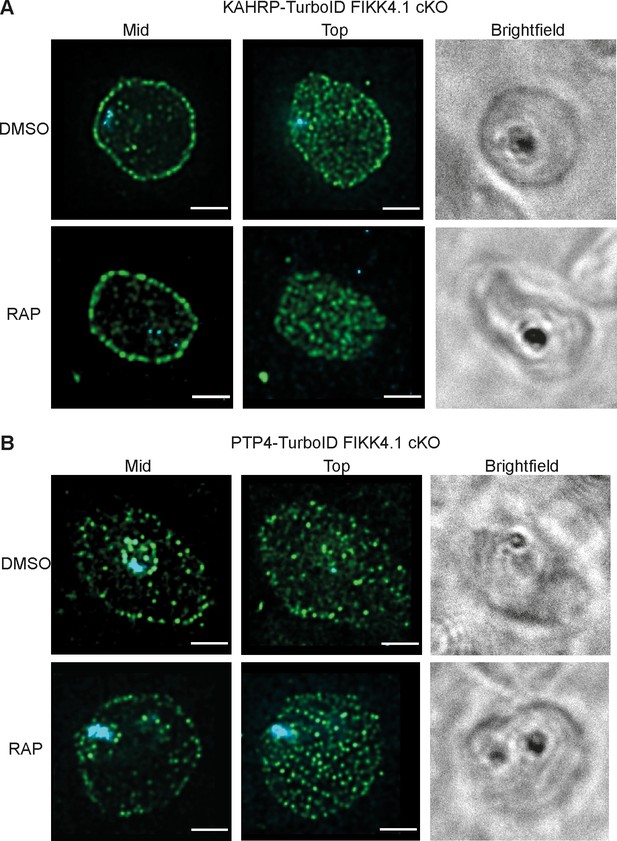
Structured illumination microscopy of KAHRP and PTP4-TurboID lines.
KAHRP-TurboID (A) and PTP4-TurboID (B) fusion lines treated with rapamycin (FIKK4.1 KO) or DMSO (Wild type), probed with anti-V5 antibody (green), and DAPI (blue). Z stacks were selected from either the center (mid) of the cell or the top, to allow the distribution of peripheral puncta to be visualized. Scale bar: 2 µm.
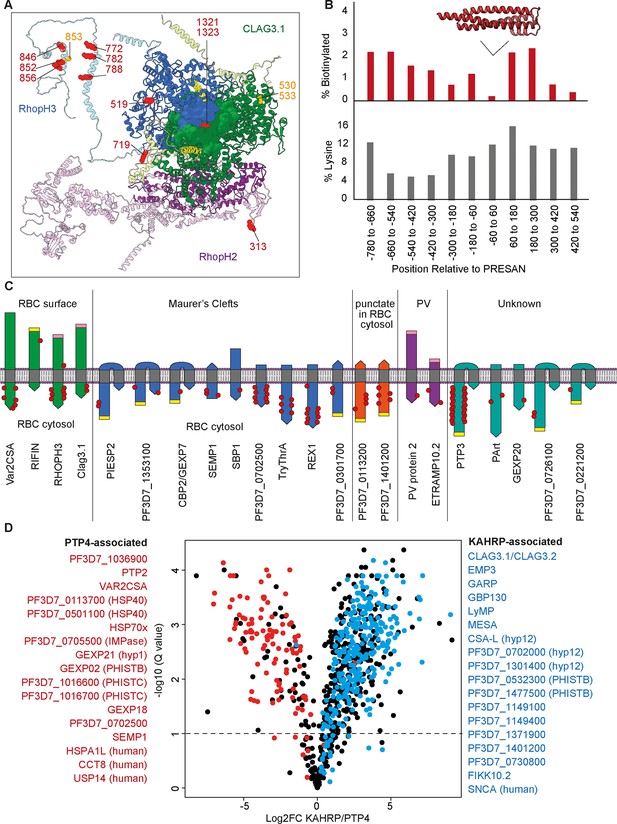
Insights into protein structure, interactions, topology, and localization from KAHRP and PTP4 site-specific TurboID.
(A) Cryo-electron microscopy structure (PDB: 7KIY) of the RHOPH complex (RHOPH2, purple; RHOPH3, blue; Clag3.1, green) (Schureck et al., 2021). Unresolved regions have been substituted with the AlphaFold-predicted structures and rotated to prevent clashes (lighter shades) (Jumper et al., 2021). Lysines biotinylated by either KAHRP or PTP4-TurboID are represented as red side-chain spheres. Residues that are either more or less phosphorylated upon FIKK4.1 deletion are in orange. Transmembrane domains are in yellow. Image created in Chimera (Pettersen et al., 2004). (B) A representation of the position of biotinylation of lysines across all 29 Plasmodium helical interspersed subtelomeric (PHIST) proteins in the dataset. Positions (x-axis) are relative to the predicted center of the ~120 aa PRESAN domain which spans positions –60 to 60 (PRESAN position predicted by PFAM via PlasmoDB; Amos et al., 2022). Top panel: y-axis represents the percentage of all lysine residues which are biotinylated in either KAHRP or PTP4 datasets. Bottom panel: percentage of lysines in primary amino acid sequence. A drop in biotinylation is observed within the PRESAN domain itself, with no reduction in the proportion of lysine residues. (C) Schematic depicting transmembrane domain (gray) proteins observed in the dataset, with the orientation based either on the literature (for PfEMP1, RIFIN, and SBP1) or predicted from the position of biotinylation sites. Some MC and punctate proteins may be trafficked in chaperoned complexes before insertion into a membrane at a later stage – we depict this hypothetical ‘membrane-inserted’ form. N-termini are flat, C-termini are pointed. PEXEL motifs, which are cleaved during export, are in yellow. Predicted TM domains and signal peptides upstream of the PEXEL motif are removed. Cleaved signal peptides with no PEXEL motif are in pink. Proteins are grouped according to published subcellular localization. Where proteins occupy more than one subcellular position (e.g. PIESP2 has been found in MC and on the surface), they are placed in one group only. The approximate positions of biotinylated residues are depicted with red dots. Proteins are not to scale. (D) Volcano plot depicting the log2 fold change in biotinylated peptide abundance between KAHRP and PTP4-TurboID fusion parasites. Proteins for which almost all peptides are more abundant in either PTP4 (red) or KAHRP (blue) datasets are highlighted. Peptides originating from KAHRP or PTP4 themselves have been removed for clarity. y-axis (-log10 q-value, N = 3).
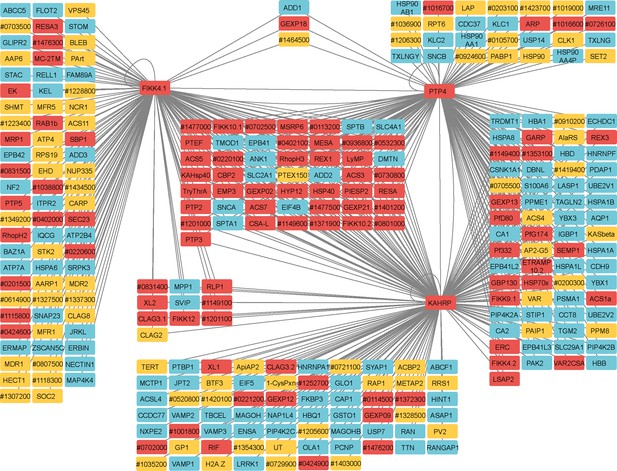
Network analysis of proteins observed in FIKK4.1, KAHRP, and PTP4 TurboID datasets.
Human proteins are in blue, P. falciparum-predicted exported proteins in red, and other P. falciparum proteins in yellow. ‘Pf3D7’ has been omitted from P. falciparum gene IDs. Connecting lines indicate a gene is likely in the vicinity of the TurboID-tagged gene. Network created with Cytoscape software (Shannon et al., 2003).
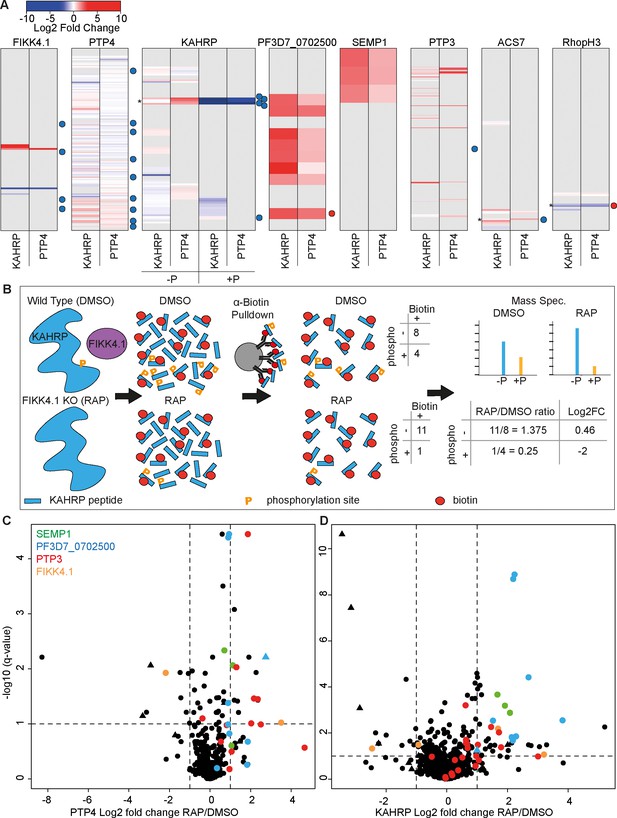
Changes in site-specific biotinylation observed upon FIKK4.1 deletion.
(A) Barcodes representing selected proteins from the PerTurboID dataset. Each row represents a lysine, with the N-terminus of the protein at the top. Lysines not biotinylated in either the KAHRP (left) or PTP4 (right) dataset are in gray. Biotinylated lysines are colored according to the log2 fold change in abundance upon FIKK4.1 deletion (RAP/DMSO), with more abundant biotinylated peptides in red and less abundant in blue. For KAHRP, peptides which are both phosphorylated and biotinylated are shown in the right-hand panels (P+) while those biotinylated only are on the left (P-). Phosphorylated peptides for all other proteins are shown in Figure 4—figure supplement 1. Dots to the right of the barcodes represent the approximate position of phosphorylation sites on the protein which were previously shown to be affected by FIKK4.1 deletion (blue, less phosphorylated; red, more phosphorylated upon FIKK4.1 deletion). Asterisks denote positions of interest (351–364 of KAHRP, 876 of ACS7, and 853 of RHOPH3). (B) Schematic demonstrating how PerTurboID can detect changes in protein phosphorylation, using KAHRP as an example. Peptides are represented as blue bars with either one or two modifications: phosphorylation (orange ‘P’) or biotin (red circle). Biotinylated peptides that are also phosphorylated are quantified separately from peptides that are only biotinylated. Loss of phosphorylated + biotinylated peptides upon FIKK4.1 deletion therefore leads to an increase in the biotinylated-only peptide and a positive log2 fold change. (C, D) Volcano plots showing the log2 fold change in biotinylated site abundance between FIKK4.1 wildtype and kinase-deleted conditions (RAP/DMSO) for PTP4-TurboID (B) and KAHRP-TurboID (C) fusion parasites. Biotinylated sites from four parasite proteins, SEMP1, PTP3, Pf3D7_0702500, and FIKK4.1, are highlighted. Sites which are also phosphorylated are represented by triangles.
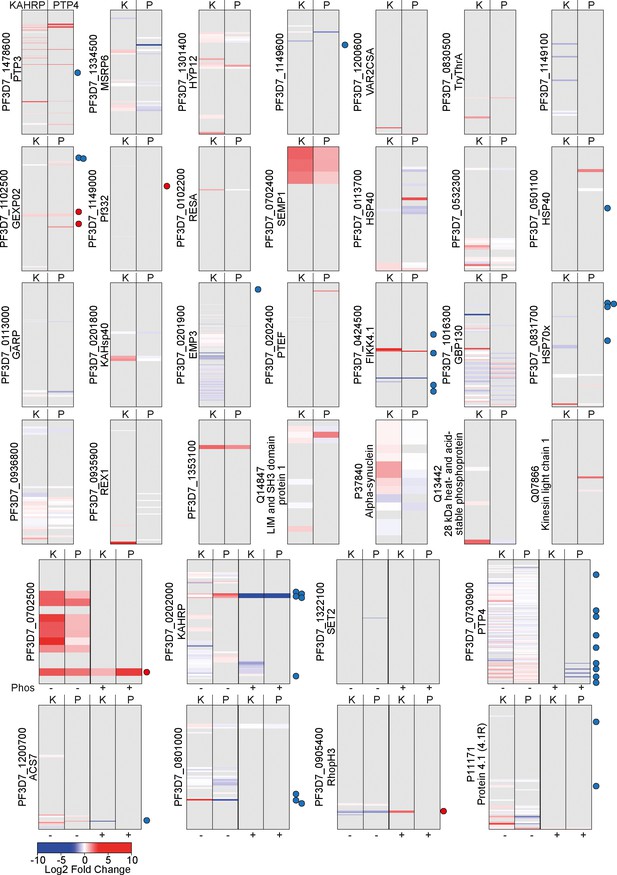
Barcodes representing changes to lysine biotinylation upon FIKK4.1 KO on proteins in the PerTurboID dataset.
Proteins for which at least two biotinylation sites were observed and at least one biotinylation site was significantly changed upon FIKK4.1 KO are included. Each row represents a lysine from the protein, with the N-terminus at the top. Lysines not biotinylated in either the KAHRP (K, left) or PTP4 (P, right) dataset are in gray. Lysines observed in the dataset are colored according to the log2 fold change in abundance upon FIKK4.1 deletion (RAP/DMSO), with peptides more abundant in the DMSO condition in blue and those more abundant in the RAP condition in red. Dots to the right of the barcodes represent the approximate position of phosphorylation sites on the protein which are affected by FIKK4.1 deletion according to our previous phosphoproteomics data (blue, less phosphorylated; red, more phosphorylated upon FIKK4.1 deletion). For the bottom eight proteins, peptides which are both phosphorylated and biotinylated are shown on the right-hand panels (P+) while those biotinylated only are on the left (P-).
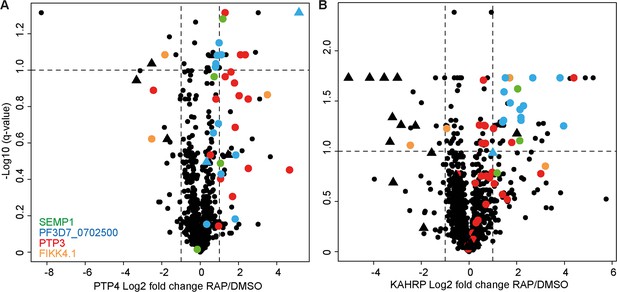
Peptide-level volcano plots.
Plots represent the log2 fold change in biotinylated peptide abundance between FIKK4.1 intact and kinase-deleted conditions (RAP/DMSO) for PTP4-TurboID (A) and KAHRP-TurboID (B) fusion parasites. Biotinylated peptides from four parasite proteins, SEMP1, PTP3, Pf3D7_0702500, and FIKK4.1, are highlighted. Peptides which are also phosphorylated are represented by triangles. Related to Figure 4C and D, where the intensities of peptides representing the same biotinylated site are averaged.
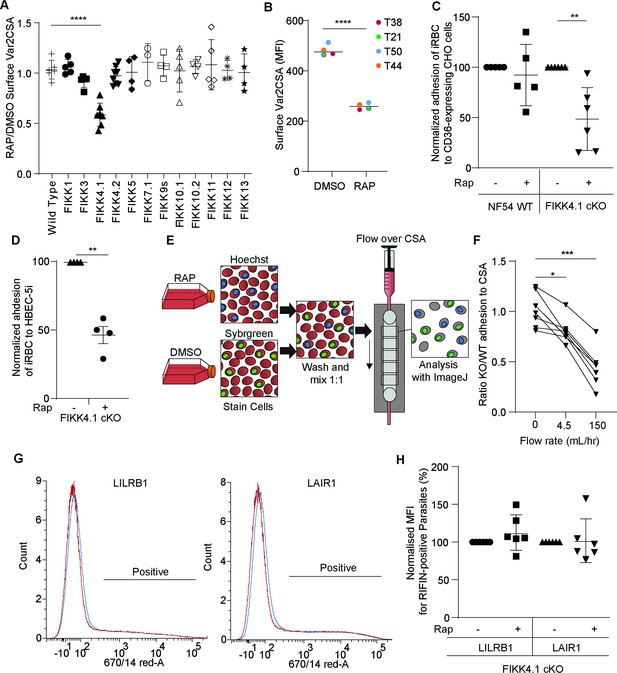
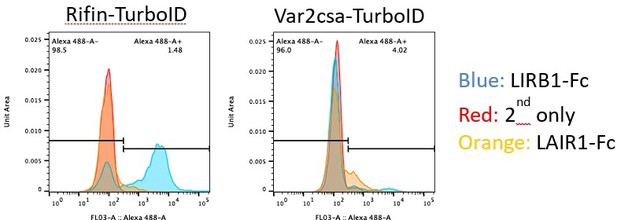
FIKK4.1 is the only FIKK kinase to modulate surface translocation of PfEMP1, but not of RIFINs.
(A) Var2CSA surface translocation analysis by flow cytometry using anti-Var2CSA antibodies (Srivastava et al., 2011). y-axis, ratio of median fluorescence intensity of Var2CSA-positive cells between RAP and DMSO-treated parasites lines. x-axis, conditional kinase domain deletion lines for the indicated FIKK kinase. ‘FIKK 9s’ represents the deletion of seven FIKK kinases on chromosome 9. Analyzed by one-way ANOVA with Dunnet’s multiple-comparison test. FIKK4.1 vs. wildtype – adjusted p-value<0.0001, n = 3–6. (B) Flow cytometry analysis of the surface presentation of Var2CSA in WT (DMSO) or FIKK4.1 KO parasites (RAP) using monoclonal antibodies directed towards different extracellular domains of the protein. y-axis, median fluorescence intensity. Paired t-test p-value<0.0001. (C) Adhesion assays with NF54 parental or FIKK4.1 cKO parasites to CD36-expressing CHO cells under flow, with or without RAP treatment, normalized to the no RAP control. Analyzed by one-way ANOVA with Sidak’s multiple-comparison adjustment. FIKK4.1 – Rap vs. +Rap adjusted p-value=0.0014. (D) Binding of iRBC to CSA-expressing HBEC-5i cells under flow. Numbers are normalized to the DMSO control. t-test p-value=0.0033. (E) Schematic depicting the set-up for semi-automated analysis of adhesion to CSA under flow. Dyed wildtype or KO parasites are premixed and flowed over CSA-coated channels. Images are taken along the channel and the ratio of wildtype to KO quantified by ImageJ at different flow speeds. (F) FIKK4.1 KO/WT adhesion under flow using the method depicted in (E). x-axis, flow rate in mL/hr. Images for ‘before’ measurements were taken with a separate uncoated channel with no flow. y-axis, ratio of WT/KO parasites at different flow speeds. Analysis by paired one-way ANOVA with corrected for multiple comparisons by the Sidak method. p-values: before vs. 4.5 mL/hr = 0.0228, before vs. 150 mL/hr = 0.0008. (G) Representative flow cytometry histograms of RIFIN presentation on FIKK4.1 WT (red) or KO (blue) infected red blood cell (iRBC). iRBCs were incubated with LILRB1-Fc or LAIR1-Fc fusion proteins, followed by anti-human Fc-APC and SYBR green staining. Parasites positive for LILRB1 or LAIR1-binding RIFINs are indicated. (H) Histograms from (G) are quantified, with the median fluorescence intensity of positive parasites normalized to the untreated control. No significant difference was observed.
Additional files
-
Supplementary file 1
Unfiltered raw data for FIKK4.1-TurboID experiment.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp1-v2.xlsx
-
Supplementary file 2
Processed FIKK4.1 TurboID data.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp2-v2.xlsx
-
Supplementary file 3
Unfiltered and unprocessed data for KAHRP and PTP4 PerTurboID experiment.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp3-v2.xlsx
-
Supplementary file 4
KAHRP and PTP4 PerTurboID processed site-level data.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp4-v2.xlsx
-
Supplementary file 5
KAHRP and PTP4 PerTurboID processed peptide-level data.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp5-v2.xlsx
-
Supplementary file 6
Network analysis of proteins observed in FIKK4.1, KAHRP, and PTP4 TurboID.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp6-v2.xlsx
-
Supplementary file 7
PHIST proteins in dataset.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp7-v2.xlsx
-
Supplementary file 8
KAHRP vs. PTP4 TurboID peptide-level data.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp8-v2.xlsx
-
Supplementary file 9
KAHRP and PTP4-TurboID phosphorylated proteins.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp9-v2.xlsx
-
Supplementary file 10
Primer list.
- https://cdn.elifesciences.org/articles/86367/elife-86367-supp10-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86367/elife-86367-mdarchecklist1-v2.docx