Improved isolation of extracellular vesicles by removal of both free proteins and lipoproteins
Figures
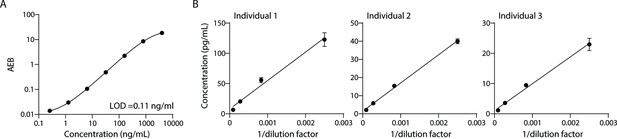
Validation of ApoB-100 Simoa assay.
Simoa ApoB-100 assay was validated using: (A) Calibration curve using purified ApoB-100 protein. (B) Dilutions of human plasma (from three different individuals) to confirm dilution linearity of endogenous ApoB-100. Error bars represent the standard deviation from two technical replicates.
-
Figure 1—source data 1
Validation of Simoa ApoB-100 assay.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig1-data1-v1.xlsx
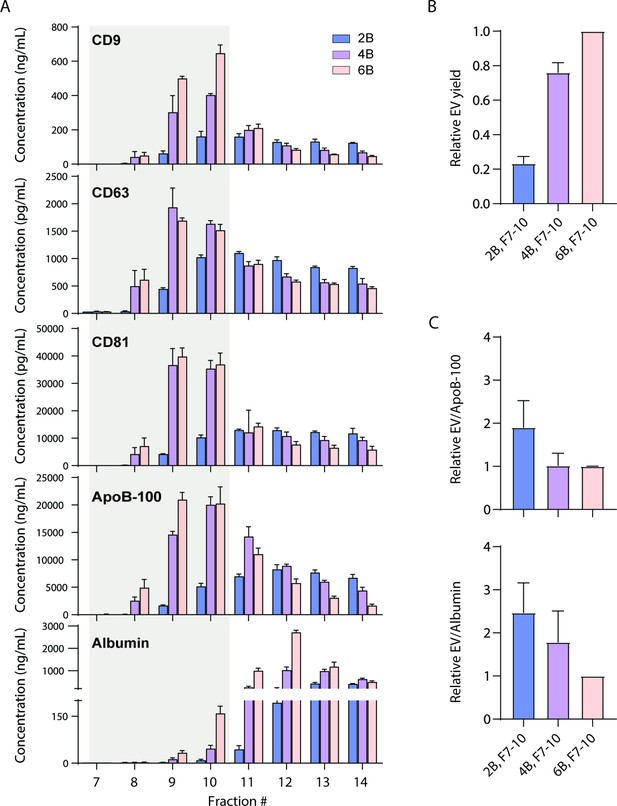
Size exclusion chromatography (SEC) of plasma using different resins.
(A) Levels of CD9, CD63, CD81, ApoB-100, and albumin were measured by Simoa after SEC of 1 ml plasma in each fraction using either Sepharose CL-2B, Sepharose CL-4B, or Sepharose CL-6B resin. (B) Extracellular vesicle (EV) yield is calculated in fractions 7–10 for Sepharose CL-2B, Sepharose CL-4B, or Sepharose CL-6B by averaging the ratios of CD9, CD63, and CD81. (C) Purity of EVs with respect to lipoproteins or free proteins is calculated by dividing relative EV yield (the average of the ratios of CD9, CD63, and CD81) by levels of ApoB-100 (top) or albumin (bottom). Error bars represent the standard deviation of four columns measured on different days with two technical replicates each.
-
Figure 2—source data 1
Simoa data (protein concentrations) for fractions of SEC columns with different resins.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Simoa data (protein concentrations) for SEC column with different number of washes.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig2-data2-v1.xlsx
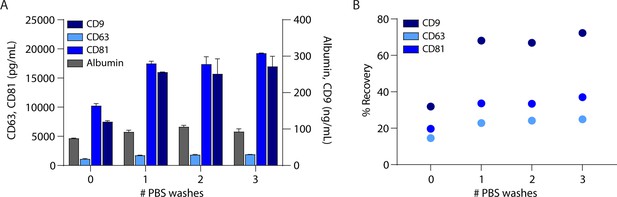
In-column phosphate-buffered saline (PBS) washes improve extracellular vesicle (EV) recovery.
(A) Levels of CD9, CD63, CD81, and albumin were measured by Simoa after EV isolation from 1 ml plasma with size exclusion chromatography (SEC) using 0, 1, 2, or 3 in-column 10 ml PBS washes. Error bars represent the standard deviation from two technical replicates. (B) Percent recovery of EVs using average of ratios of CD9, CD63, and CD81 in SEC isolation relative to plasma.
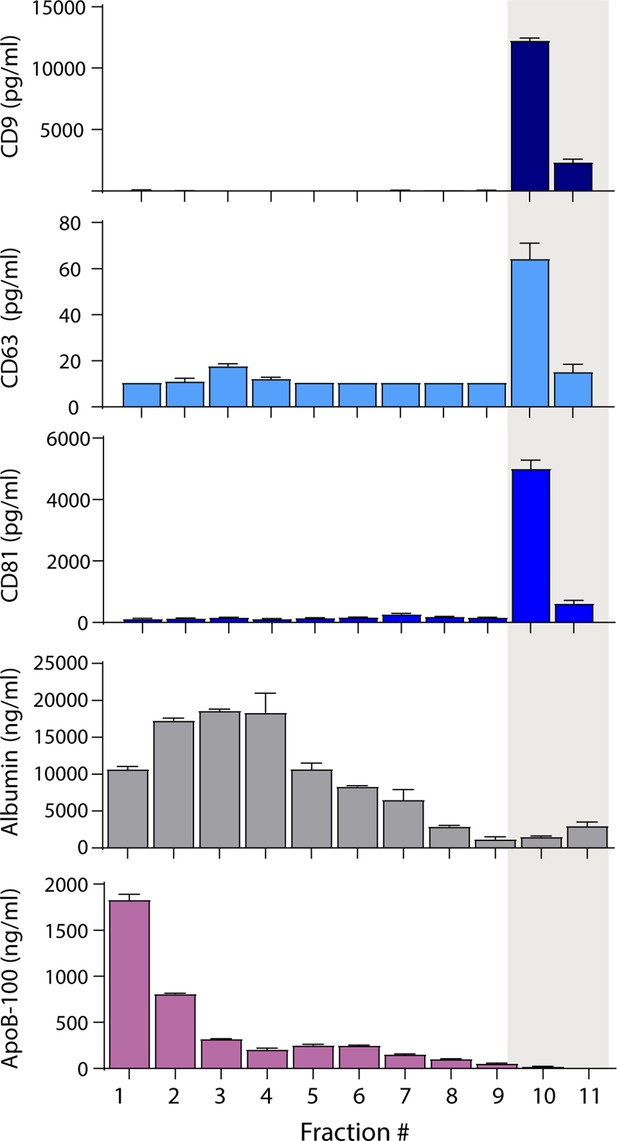
Separation of extracellular vesicles (EVs), lipoproteins, and free proteins from plasma using density gradient centrifugation.
Levels of CD9, CD63, CD81, albumin, and ApoB-100 were measured by Simoa in individual 1 ml fractions (collected from the top) after density gradient centrifugation of plasma using an iodixanol gradient. Error bars represent the standard deviation of two replicates of each measurement.
-
Figure 3—source data 1
Simoa data (protein concentrations) for different density gradient centrifugation fractions.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig3-data1-v1.xlsx
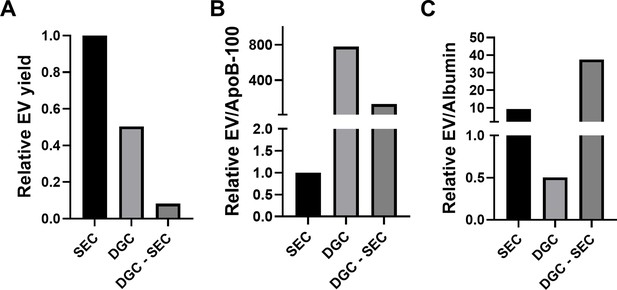
Comparison of density gradient centrifugation to size exclusion chromatography (SEC).
Levels of CD9, CD63, CD81, ApoB-100 albumin were measured (in duplicate and then averaged) by Simoa to compare extracellular vesicle (EV) isolation from 1 ml plasma using density gradient (DG) centrifugation, SEC, or density gradient centrifugation followed by size exclusion chromatography (DG-SEC). For the DG and DG-SEC condition, fraction 10 was analyzed. Simoa measurements were used to quantify relative EV recovery (A), EV/ApoB-100 ratio (B), and EV/albumin ratio (C).
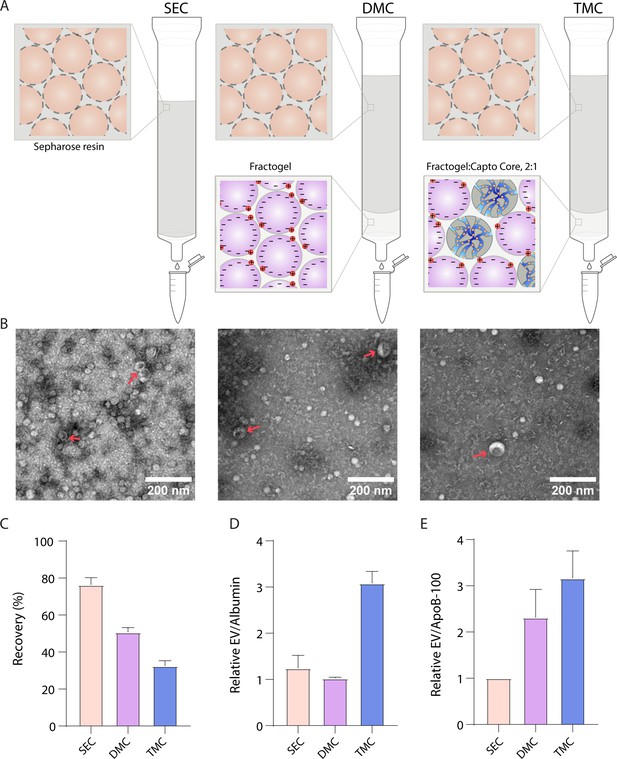
Comparison of novel columns for extracellular vesicle (EV) isolation from plasma using electron microscopy and Simoa.
(A) Schematic of the columns being compared: size exclusion chromatography (SEC) column comprised of 10 ml Sepharose CL-6B, dual-mode chromatography (DMC) columns comprised of 10 ml Sepharose CL-6B SEC resin atop 2 ml Fractogel cation exchange resin, Tri-Mode Chromatography (TMC) columns comprised of 10 ml Sepharose CL-6B SEC resin atop 2 ml 2:1 ratio of 2 ml Fractogel cation exchange resin to Capto Core 700 multimodal chromatography resin. (B) Electron microscopy of EVs isolated from plasma using SEC (left), DMC (middle), or TMC (right) columns. EVs indicated with red arrows (among background of lipoproteins). (C) EV recovery is calculated for EV isolation from plasma for SEC (fractions 7–10), DMC (fractions 9–12), or TMC (fractions 9–12). Simoa measurements in the designated fractions for CD9, CD63, and CD81 are taken as a ratio relative to measurements of these proteins from diluted plasma and these three ratios are then averaged to calculate recovery. (D) Purity of EVs with respect to free proteins is determined by dividing relative EV yield (the average of the ratios of CD9, CD63, and CD81) by relative levels of albumin in each condition. (E) Purity of EVs with respect to lipoproteins is determined by dividing relative EV yield (the average of the ratios of CD9, CD63, and CD81) by relative levels of ApoB-100 in each condition. Error bars represent the standard deviation of four column measured on different days with two technical replicates each.
-
Figure 4—source data 1
Simoa data (protein concentrations) for TMC columns with different ratios of resins in the bottom layer.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Simoa data (protein concentrations) for different fractions of SEC and DMC columns.
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Simoa data (protein concentrations) comparing SEC (fractions 7-10), DMC and TMC columns (fractions 9-12).
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig4-data3-v1.xlsx
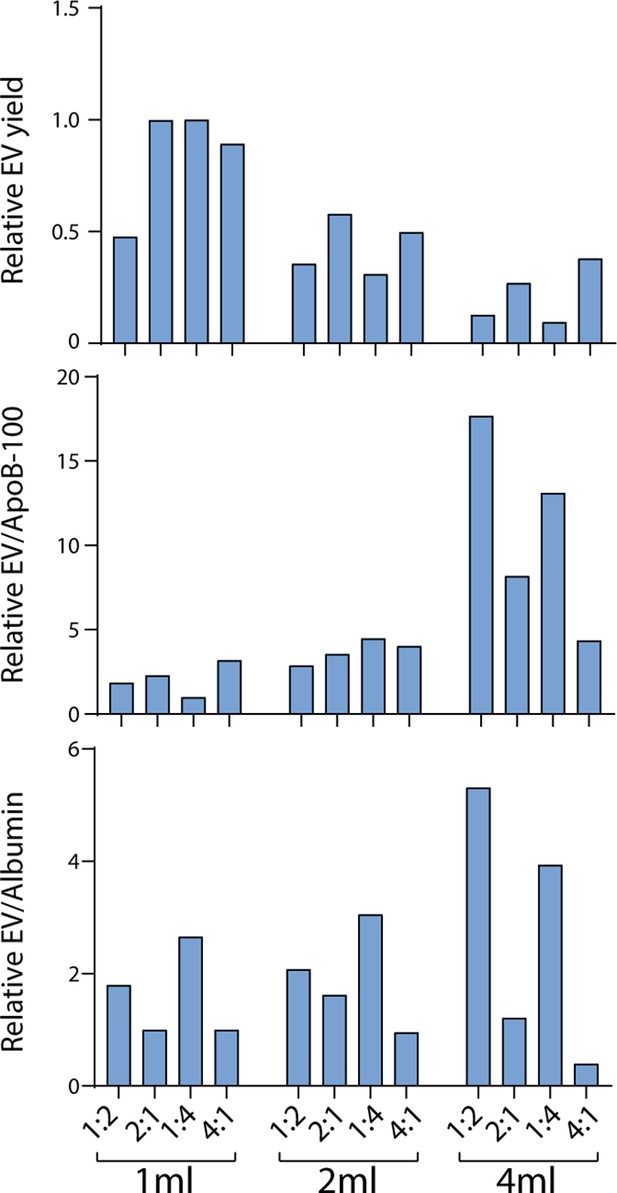
Comparison of resin volumes and ratios for Tri-Mode Chromatography (TMC) column.
Levels of CD9, CD63, CD81, ApoB-100, and albumin were measured (in duplicate and then averaged) by Simoa in extracellular vesicle (EV) samples isolated from 1 ml plasma to compare TMC columns with different volumes and ratios of Fractogel cation exchange resin to Capto Core 700 resin. All conditions describe the bottom layer under a 10 ml Sepharose CL-6B top layer. The 1, 2, or 4 ml volume of the bottom later indicates the volume of the solid resin mixture of Fractogel cation exchange resin and Capto Core 700 resin. The following fractions were collected for each: 8–11 for 1 ml bottom layer, 9–12 for 2 ml bottom layer, and 11–14 for 4 ml bottom layer.
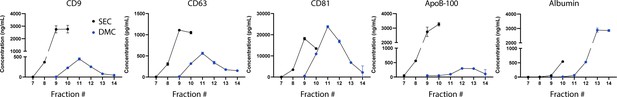
Analysis of markers in individual fractions of size exclusion chromatography (SEC) and dual-mode chromatography (DMC).
Levels of CD9, CD63, CD81, ApoB-100, and albumin were measured by Simoa in fractions 7–10 for SEC with 10 ml Sepharose CL-6B column and fractions 7–14 for DMC using a column with 2 ml Fractogel cation exchange bottom layer and 10 ml Sepharose CL-6B top layer. Error bars represent the standard deviation from two technical replicates.
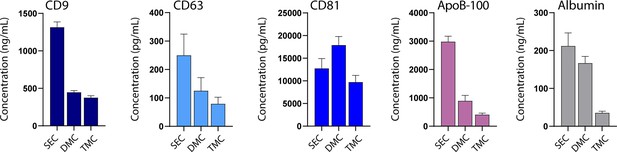
Comparison of marker levels in size exclusion chromatography (SEC), dual-mode chromatography (DMC), and Tri-Mode Chromatography (TMC).
Levels of CD9, CD63, CD81, ApoB-100, and albumin were measured by Simoa in extracellular vesicle (EV) samples isolated from 1 ml plasma using SEC (fractions 7–10), DMC (fractions 9–12), or TMC columns (fractions 9–12). Error bars represent the standard deviation of four column measured on different days with two technical replicates each.
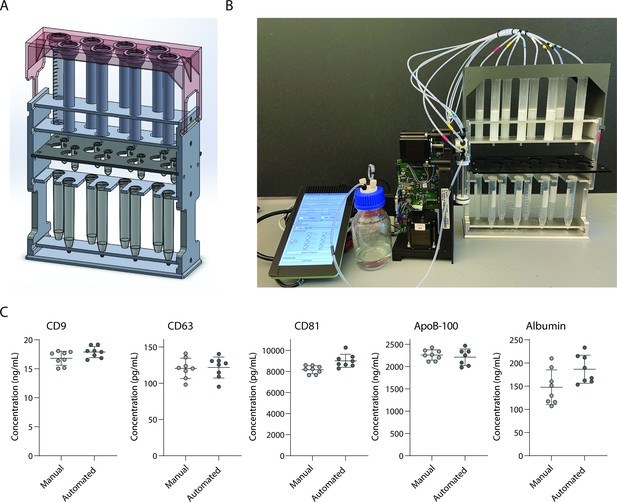
Development and validation of automated device for running size exclusion chromatography (SEC) columns in parallel.
(A) CAD image of semi-automated SEC stand designed to hold eight columns at once with sliding collection tube holder that allows liquid to drip either into 2 ml collection tubes, or to waste. (B) Photograph of stand connected to a Tecan Cavro syringe pump controlled by a Raspberry Pi. (C) Simoa comparison of CD9, CD63, CD81, ApoB-100, and albumin when SEC was performed on 16 samples of 1 ml plasma using either manual SEC (8 samples) or SEC on the automated device (8 samples). Each point is the average of two Simoa measurements (technical replicates).
-
Figure 5—source data 1
Simoa data (protein concentrations) comparing manual and automated SEC EV isolation (fractions 7-10).
- https://cdn.elifesciences.org/articles/86394/elife-86394-fig5-data1-v1.xlsx
Tables
Spike and recovery for ApoB-100 assay.
Percent recovery of different concentrations of purified ApoB-100 spike added to plasma and measured using the ApoB-100 Simoa assay.
| Spike concentration(ng/ml) | Average recovery (%) | |
|---|---|---|
| Spike 1 | 5 | 94.7 |
| Spike 2 | 10 | 87.2 |
| Spike 3 | 50 | 90.8 |
| Spike 4 | 500 | 90.6 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (human) | Plasma | BioIVT | Cat# HUMANPLK2PNN | Pooled gender, K2EDTA |
| Antibody | anti-CD9 (rabbit monoclonal) | Abcam | Cat# ab263024 | Simoa capture (0.031 μg per assay) |
| Antibody | anti-CD9 (mouse monoclonal) | Abcam | Cat# ab58989 | Simoa detector (0.06 μg per assay) |
| Antibody | anti-CD63 (mouse monoclonal) | R&D Systems | Cat# MAB5048 | Simoa capture (0.031 μg per assay) |
| Antibody | anti-CD63 (mouse monoclonal) | BD | Cat# 556019 RRID: AB_396297 | Simoa detector (0.0435 μg per assay) |
| Antibody | anti-CD81 (mouse monoclonal) | Abcam | Cat# ab79559 | Simoa capture (0.031 μg per assay) |
| Antibody | anti-CD81 (mouse monoclonal) | BioLegend | Cat# 349502 RRID: AB_10643417 | Simoa detector (0.0435 μg per assay) |
| Antibody | Human Serum Albumin DuoSet ELISA | R&D Systems | Cat# DY1455 | Simoa capture (0.031 μg per assay) and detector (0.002 μg per assay) |
| Antibody | anti-ApoB (mouse monoclonal) | R&D Systems | Cat# mab4124 RRID:AB_2057095 | Simoa capture (0.031 μg per assay) |
| Antibody | anti-ApoB (mouse monoclonal) | R&D Systems | Cat# mab41242 | Simoa detector (0.08 μg per assay) |
| Peptide, recombinant protein | CD9 | Abcam | Cat# ab152262 | |
| Peptide, recombinant protein | CD63 | Origene | Cat# TP301733 | |
| Peptide, recombinant protein | CD81 | Origene | Cat# TP317508 | |
| Peptide, recombinant protein | Albumin | Abcam | Cat# ab201876 | |
| Other | Purified ApoB-100 Standard | Origene | Cat# BA1030 | Protein standard for Simoa |
| Other | Sepharose CL-2B | Cytiva | Cat# 17014001 | Resin for SEC |
| Other | Sepharose CL-4B | Cytiva | Cat# 17015001 | Resin for SEC |
| Other | Sepharose CL-6B | Cytiva | Cat# 17016001 | Resin for SEC |
| Other | Fractogel EMD SO3- (M) | MilliporeSigma | Cat# 1168820100 | Resin for DMC/TMC |
| Other | Capto Core 700 multimodal chromatography resin | Cytiva | Cat# 17548102 | Resin for TMC |
| Other | Econo-Pac Chromatography Columns | Bio-Rad | Cat # 7321010 | Empty columns |
Additional files
-
Supplementary file 1
Protein list identified by mass spectrometry of extracellular vesicles (EVs) isolated from plasma using Tri-Mode Chromatography (TMC).
- https://cdn.elifesciences.org/articles/86394/elife-86394-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86394/elife-86394-mdarchecklist1-v1.pdf





