CRB3 navigates Rab11 trafficking vesicles to promote γTuRC assembly during ciliogenesis
Figures
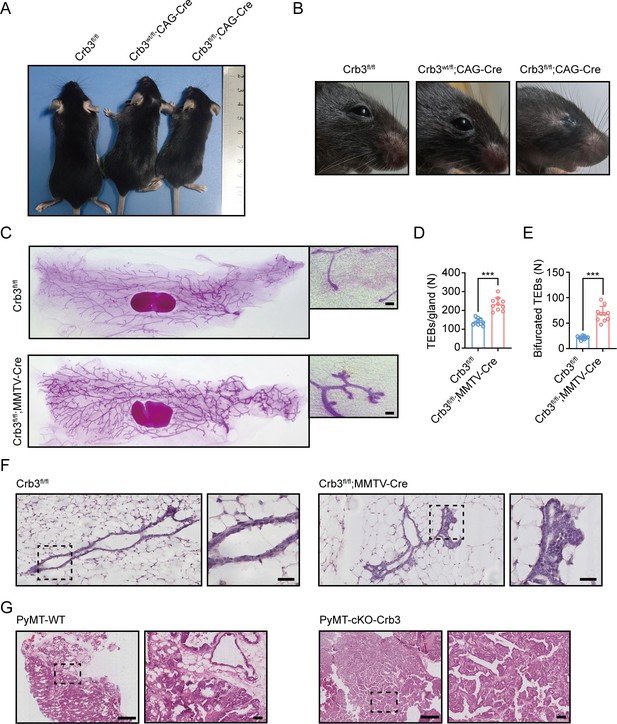
Crb3 knockout mice exhibit smaller sizes and ocular abnormalities, and mammary epithelial cell-specific Crb3 knockout leads to ductal epithelial hyperplasia and promotes tumorigenesis.
(A, B) Representative whole bodies (A) and eyes (B) from littermate Crb3fl/fl, Crb3wt/fl;CAG-Cre and Crb3fl/fl;CAG-Cre mice at 4 weeks old. (C) Representative mammary whole mounts from littermate Crb3fl/fl and Crb3fl/fl;MMTV-Cre mice at 8 weeks old with Carmine-alum staining. (scale bars, 200 μm) (D, E) Quantification of the average number of terminal end buds (TEBs) and bifurcated TEBs in littermate Crb3fl/fl (n = 10) and Crb3fl/fl;MMTV-Cre (n = 10) mice at 8 weeks old. (F) Representative images of mammary glands in littermate Crb3fl/fl and Crb3fl/fl;MMTV-Cre mice stained with H&E (scale bars, 50 μm). (G) Representative images of primary tumors stained with H&E in PyMT-WT and PyMT-cKO-Crb3 mice at 9 weeks old (scale bars, left 500 μm, right 50 μm). The magnified areas of boxed sections are shown in the right panels. Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
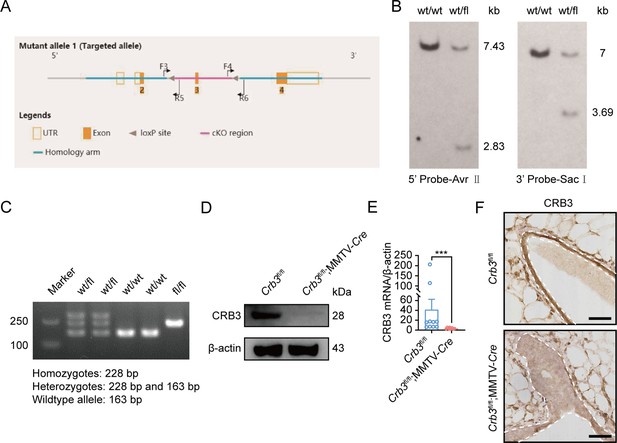
Generation of Crb3 knockout mice using the Cre-loxP system.
(A) Schematic representation of the strategy for loxP sequence insertion and PCR genotyping. The loxP sites (gray arrow) flanked either side of exon 3 (orange) in the Crb3 gene. The F3&R5 and F4&R6 primer pair were used to differentiate the alleles. (B) Southern blot analysis of a positive embryonic stem (ES) cell clone confirmed homologous recombination. The genomic DNA from ES cell clones and wild-type cells was digested with Avr II or Sac I and hybridized with a 5′ external probe or a 3′ internal probe. (C) PCR identification of mouse genotyping showing Crb3wt/wt, Crb3wt/fl, and Crb3fl/fl alleles. (D) The expression of Crb3 was detected by immunoblotting in mammary gland tissues in Crb3fl/fl and Crb3fl/fl;MMTV-Cre mice at 8 weeks old. (E) Real-time quantitative PCR showing the relative mRNA expression of Crb3 in mammary gland tissues (n = 10 mice). (F) Crb3 expression was assessed using immunohistochemistry in mammary gland tissues. The mammary epithelial cells are marked by areas of white dotted lines (scale bars, 50 μm). Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
-
Figure 1—figure supplement 1—source data 1
Original gel or blot images of Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig1-figsupp1-data1-v1.zip
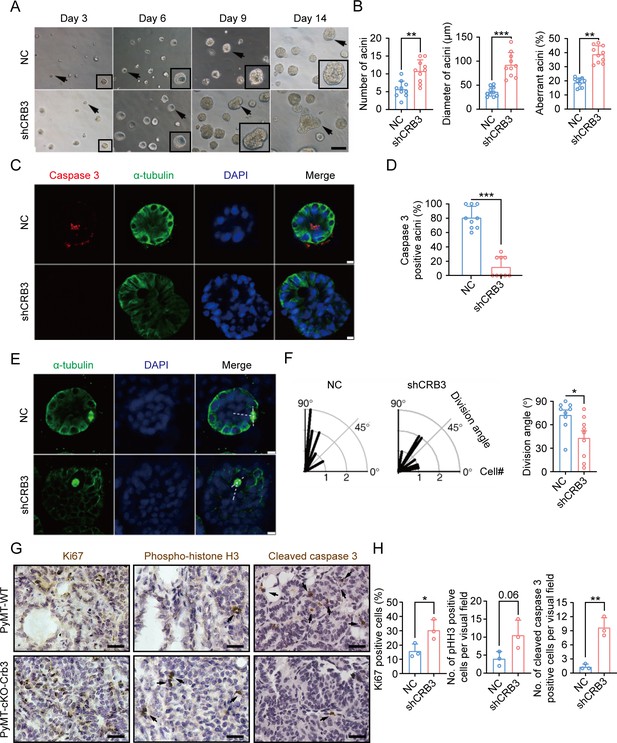
CRB3 knockdown inhibits acinar formation of mammary epithelial cells in a 3D culture system.
(A) Representative effect of CRB3 on acinar formation in the 3D culture system at days 3, 6, 9, and 14. (The magnified areas of the marked arrows are shown in the lower-right corner.) (B) Quantification of the average number, diameter, and aberration of acini (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). (C) Immunofluorescence showing apoptosis during lumen formation. Caspase 3 (red), α-tubulin (green), and DNA (blue). (D) Quantification of the proportion of caspase 3-positive acini (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments). (E) Immunofluorescence showing the mitotic spindle orientation during lumen formation. α-tubulin (green), and DNA (blue). (F) Quantification of division angle (at least 3–4 acini were randomly examined for each condition in each of three independent experiments). (G) Immunohistochemical analyses of Ki67, phospho-histone H3, and cleaved caspase 3 in primary tumors from PyMT-WT (n = 3) and PyMT-cKO-Crb3 (n = 3) mice at 9 weeks old. (Positive cells are marked by arrows; scale bars, 25 μm.) (H) Quantification of the number of Ki67, phospho-histone H3, and cleaved caspase 3-positive cells from IHC staining images. Bars represent means ± SD; unpaired Student’s t-test, *p<0.5, **p<0.01, ***p<0.001.
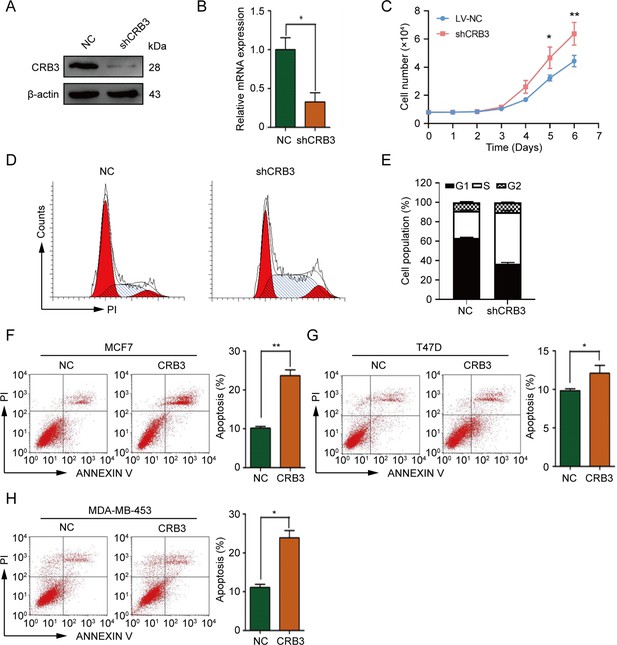
CRB3 knockdown promotes proliferation of mammary epithelial cells, while overexpression promotes apoptosis of breast cancer.
(A, B) Immunoblot and real-time quantitative PCR analysis of CRB3 knockout efficiency in MCF10A cells. (C) MCF10A proliferation assay over six successive days. (D) Cell cycle showing the distribution of MCF10A cells. (E) Quantification of cell cycle distribution. (F–H) Apoptosis of MCF7, T47D, and MDA-MB-453 breast cancer cells with CRB3b overexpression and quantification of apoptotic cells. The data are three independent experiments performed in triplicate. Bars represent means ± SD; unpaired Student’s t-test, *p<0.05, **p<0.01.
-
Figure 2—figure supplement 1—source data 1
Original blot images of Figure 2—figure supplement 1A.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig2-figsupp1-data1-v1.zip
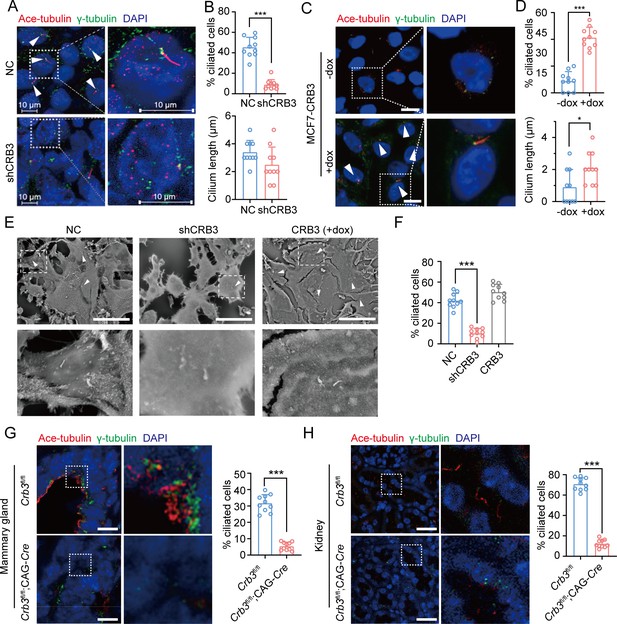
CRB3 alters primary cilium formation in mammary cells, mammary ductal lumen, and renal tubule from Crb3fl/fl;CAG-Cre mice.
(A, C) Representative images of immunofluorescent staining of primary cilium formation with CRB3 knockdown in MCF10A cells and CRB3b conditional overexpression upon dox induction in MCF7 cells. Acetylated tubulin (red), γ-tubulin (green), and DNA (blue). (The primary cilium is marked by arrows; scale bars, 10 μm.) (B, D) Quantification of the proportion of cells with primary cilium formation and the length of the primary cilium (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). (E) Representative scanning electron microscope images of primary cilium formation with CRB3 knockdown and CRB3b conditional overexpression in MCF10A cells. (The primary cilium is marked by arrows; scale bars, 50 μm.) (F) Quantification of the proportion of cells with primary cilium formation (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). (G, H) Representative immunofluorescent staining of primary cilium formation in the mammary ductal lumen and renal tubule from Crb3fl/fl (n = 10) and Crb3fl/fl;CAG-Cre (n = 10) mice, respectively. Acetylated tubulin (red), γ-tubulin (green), and DNA (blue) (scale bars, 25 μm). Bars represent means ± SD; unpaired Student’s t-test, *p<0.05, ***p<0.001.
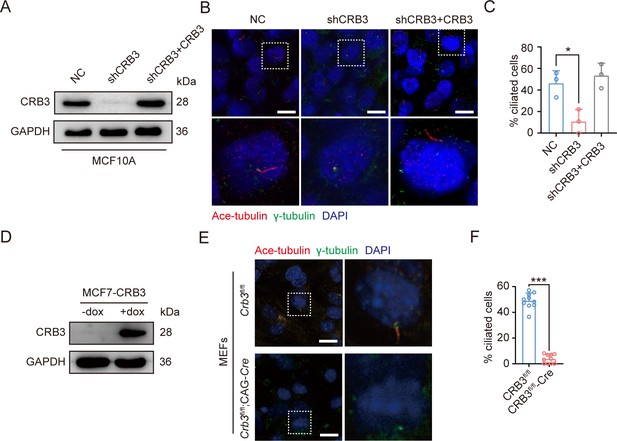
CRB3 alters primary cilium formation in MCF10A cells and mouse embryonic fibroblasts (MEFs) from Crb3fl/fl;CAG-Cre mice.
(A) Immunoblot analysis of CRB3b overexpression efficiency in CRB3-depleted MCF10A cells. (B). Immunoblot analysis of CRB3b overexpression efficiency in CRB3-depleted MCF10A cells. (C) Quantification of the proportion of cells with primary cilium formation (three random micrographs were analyzed). (D) Immunoblot analysis of CRB3b conditional overexpression upon dox induction efficiency in MCF7 cells. (E) Representative immunofluorescent staining of primary cilium formation in MEFs from Crb3fl/fl and Crb3fl/fl;CAG-Cre mice. (F) Quantification of the proportion of cells with primary cilium formation (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). Acetylated tubulin (red), γ-tubulin (green), and DNA (blue) (scale bars, 10 μm). Bars represent means ± SD; unpaired Student’s t-test, *p<0.05, ***p<0.001.
-
Figure 3—figure supplement 1—source data 1
Original blot images of Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig3-figsupp1-data1-v1.zip

CRB3 localizes to the basal body of the primary cilium.
(A) Immunofluorescence showing the co-localization of exogenous CRB3b with centrosomes in MCF10A cells. Pericentrin, a marker of centrosome (red), CRB3-GFP (green), and DNA (blue) (co-localization is marked by arrows; scale bars, 10 μm). (B) Quantification of the proportion of cells with pericentrin and exogenous CRB3b co-localization (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). (C) Another co-localization of endogenous CRB3 with the basal body in MCF10A cells. γ-Tubulin is a marker of the centrosome and basal body of the primary cilium (green), CRB3 (red), and DNA (blue) (co-localization is marked by arrows; scale bars, 10 μm). (D) Corresponding fluorescence intensity profile across a section of the array, as indicated by the dashed white line in (C). (E) Quantification of the number of γ-tubulin foci in MCF10A cells from IF staining images (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). (F) Double immunostaining displaying the co-localization of CRB3 with the primary cilium in MCF10A cells. Acetylated tubulin (green), CRB3 (red), and DNA (blue) (co-localization is marked by arrows; scale bars, 10 μm). (G) Fluorescence 3D reconstruction of CRB3 and primary cilium co-localization. Acetylated tubulin (red), CRB3 (green), and DNA (blue). Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
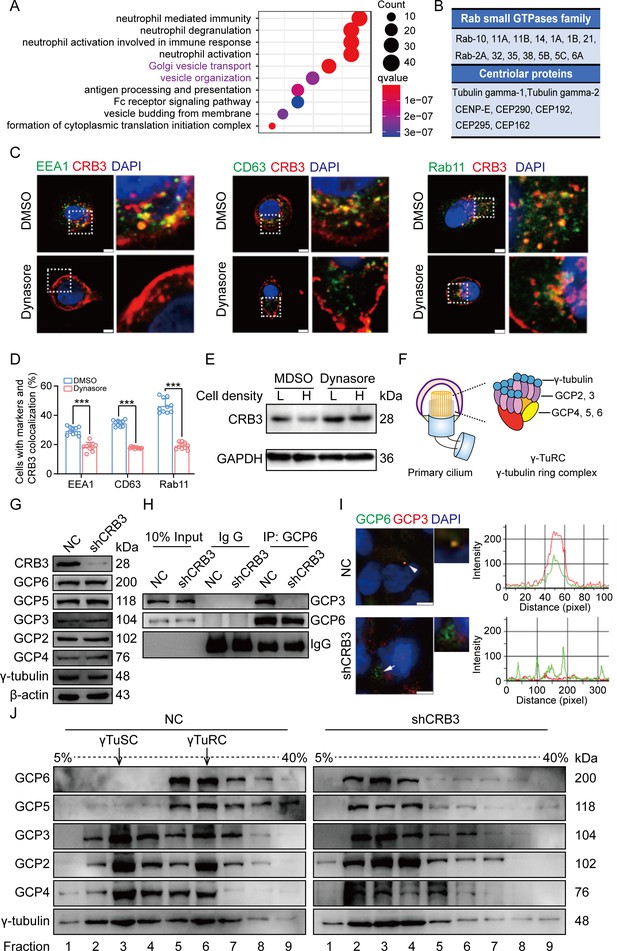
CRB3 trafficking is mediated by Rab11-positive endosomes, and CRB3 knockdown destabilizes γ-tubulin ring complex (γTuRC) assembly during ciliogenesis.
(A) Pathway aggregation analysis of CRB3b-binding proteins identified by mass spectrometry in MCF10A cells. (B) Table of some Rab small GTPase family members and centriolar proteins identified as CRB3b-binding proteins. (C) Immunofluorescence showing the co-localization of CRB3 with EEA1-, CD63-, and Rab11-positive endosomes in MCF10A cells. EEA1, CD63, Rab11 (green), CRB3 (red), and DNA (blue) (scale bars, 10 μm). (D) Quantification of the proportion of cells with these markers and CRB3 co-localization (n = 10). (E) Western blotting showing the levels of CRB3 in MCF10A cells treated with dynasore at different cell densities. (F) The structure diagram of γTuRC. (G) Immunoblot analysis of the effect of CRB3 on γTuRC molecules in MCF10A cells. (H) Coimmunoprecipitation showing the interacting proteins with GCP6 in MCF10A cells with CRB3 knockdown. (I) Representative images of immunofluorescent staining of GCP3 and GCP6 co-localization in MCF10A cells with the corresponding fluorescence intensity profile. GCP3 (red), GCP6 (green), and DNA (blue) (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, scale bars, 10 μm). (J) The comparison of cytoplasmic extracts from MCF10A cells and cells with CRB3 knockdown after fractionation in sucrose gradients. The γ-tubulin small complex (γTuSC) sedimentation was mainly in fractions 3, and γTuRC sedimentation was mainly in fractions 6. Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
-
Figure 5—source data 1
Raw data for the identification of CRB3b-binding proteins by tandem mass spectrometry related to Figure 5A and B.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Original blot images of Figure 5.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig5-data2-v1.zip
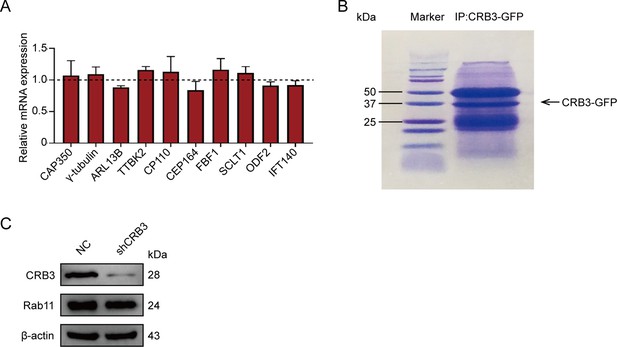
The effect of CRB3 on the expression of some ciliogenesis-related genes and Rab11.
(A) Quantification of mRNA expression of ciliogenesis-related genes in MCF10A cells (normalized to GAPDH, data are three independent experiments performed in triplicate). (B) Coomassie blue staining showing the gel electrophoresis of exogenous CRB3b immunoprecipitated precipitates. (C) Immunoblot analysis of the effect of CRB3 on Rab11 in MCF10A cells. Bars represent means ± SD; unpaired Student’s t-test.
-
Figure 5—figure supplement 1—source data 1
Original gel or blot images of Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig5-figsupp1-data1-v1.zip
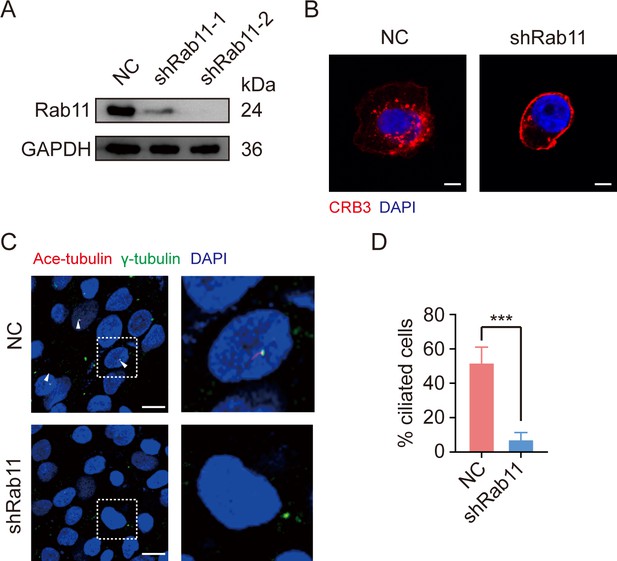
Rab11 knockdown caused alterations in CRB3 trafficking and defects in primary cilium.
(A) Immunoblot analysis of Rab11 knockout efficiency in MCF10A cells. (B) Representative images of immunofluorescent staining of CRB3 trafficking with Rab11 knockdown in MCF10A cells. CRB3 (red) and DNA (blue) (scale bars, 10 μm). (C) Representative images of immunofluorescent staining of primary cilium formation with Rab11 knockdown in MCF10A cells. Acetylated tubulin (red), γ-tubulin (green), and DNA (blue) (primary cilium is marked by arrows; scale bars, 20 μm). (D) Quantification of the proportion of cells with primary cilium formation in MCF10A cells (at least 3–4 random micrographs were analyzed for each condition in each of three independent experiments, n = 10). Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
-
Figure 5—figure supplement 2—source data 1
Original blot images of Figure 5—figure supplement 2A.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig5-figsupp2-data1-v1.zip
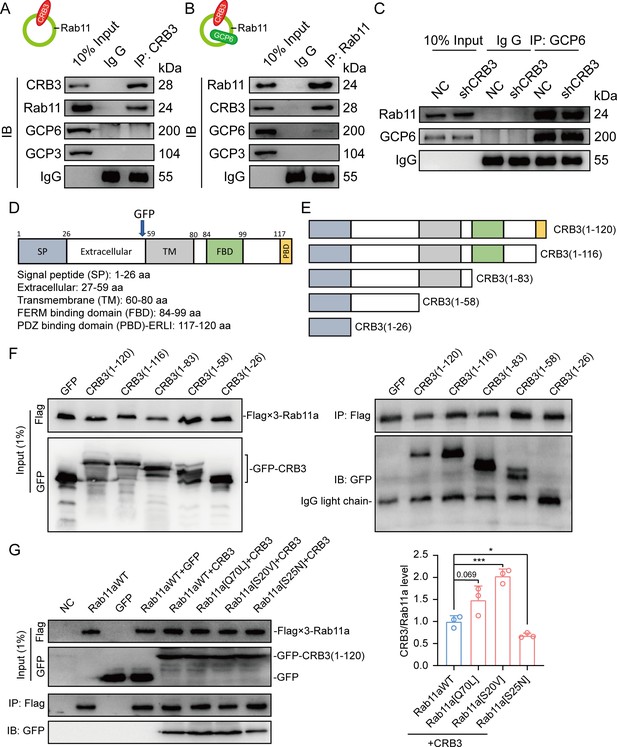
CRB3 interacts with Rab11.
(A) Coimmunoprecipitation of CRB3 with Rab11, GCP6, and GCP3 in MCF10A cells. (B) Coimmunoprecipitation of Rab11 with CRB3, GCP6, and GCP3 in MCF10A cells. (C) Coimmunoprecipitation of Rab11 with GCP6 in control and CRB3 knockdown MCF10A cells. (D) Schematic diagram of CRB3b domains. (E) Diagram truncations of CRB3b-GFP fusion proteins with serial C-terminal deletions. (F) Domain mapping of CRB3b-GFP for Flag-Rab11a binding. Flag antibody co-IP of the full-length CRB3b-GFP and truncations of CRB3b-GFP with Flag-Rab11a were cotransfected into HEK293 cells for 48 hr. Immunoblot analysis was performed using GFP and Flag antibodies. (G) Coimmunoprecipitation of Rab11 mutant variants with full-length CRB3b-GFP. Flag antibody co-IP of the full-length CRB3b-GFP with Flag-Rab11aWT, Flag-Rab11a[Q70L], Flag-Rab11a[S20V], and Flag-Rab11a[S25N] were cotransfected into HEK293 cells for 48 hr. Immunoblot analysis was performed using GFP and Flag antibodies. Bars represent means ± SD, and the experiments were performed in triplicate; unpaired Student’s t-test, *p<0.05, ***p<0.001.
-
Figure 6—source data 1
Original blot images of Figure 6.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig6-data1-v1.zip
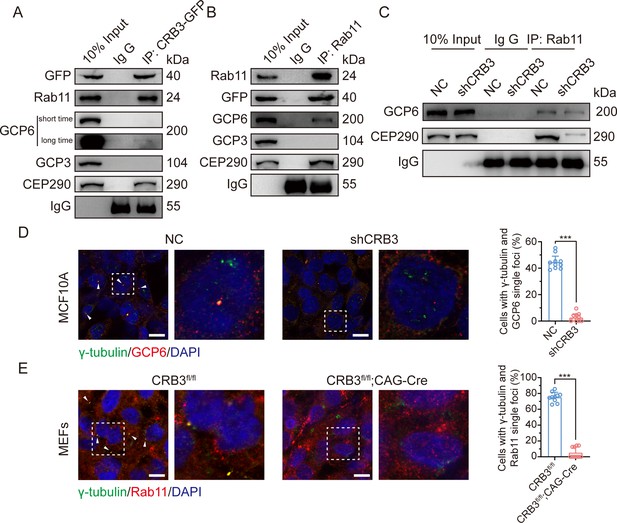
CRB3 navigates GCP6/Rab11 trafficking vesicles to the basal body of the primary cilium.
(A) Coimmunoprecipitation of exogenous CRB3b with Rab11, GCP6, GCP3, and CEP290 in MCF10A cells. (B) Coimmunoprecipitation of Rab11 with exogenous CRB3b, GCP6, GCP3, and CEP290 in MCF10A cells. (C) Coimmunoprecipitation of Rab11 with GCP6 and CEP290 in control and CRB3 knockdown MCF10A cells. (D) Representative immunofluorescent images of GCP6 and γ-tubulin co-localization of the basal body foci in MCF10A cells with CRB3 knockdown. γ-Tubulin (green), GCP6 (red), and DNA (blue) (co-localization is marked by arrows; scale bars, 25 μm). (E) Representative immunofluorescent images of Rab11 and γ-tubulin co-localization of the basal body foci in mouse embryonic fibroblast (MEF) cells from Crb3fl/fl and Crb3fl/fl;CAG-Cre mice. γ-Tubulin (green), Rab11 (red), and DNA (blue). Foci are marked by arrows; scale bars, 25 μm. Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
-
Figure 7—source data 1
Original blot images of Figure 7.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig7-data1-v1.zip

Defects in CRB3 expression inhibit ciliary assembly in breast cancer tissues and activate the Wnt signaling pathway in mammary cells and PyMT mouse model.
(A) Representative immunofluorescent images of CRB3 in breast cancer tissues (n = 50). CRB3 (green) and DNA (blue) (scale bars, 25 μm). (B) Representative immunofluorescent images of the primary cilium in breast cancer tissues (n = 50). Acetylated tubulin (red), γ-tubulin (green), and DNA (blue) (scale bars, 10 μm). (C) Quantification of the proportion of cells with primary cilium formation in breast cancer tissues (n = 50). (D) Representative immunofluorescent images of CRB3 and γ-tubulin co-localization in adjacent paracarcinoma tissues. CRB3 (green), γ-tubulin (red), and DNA (blue) (scale bars, 25 μm). (E) Quantification of the proportion of MFC10A cells with SMO translocation after CRB3 knockdown (n = 6). (F) Real-time quantitative PCR showing the relative mRNA expression of GLI1 upon SAG treatment in CRB3-depleted MFC10A cells (n = 6). (G) Immunoblot analyses of the effect of CRB3 on the molecules of the Wnt signaling pathway in mammary cells, and the experiments were performed in triplicate. (H. I) Immunohistochemical analyses of GLI1 and β-catenin in primary tumors from PyMT-WT and PyMT-cKO-Crb3 mice at 9 weeks old, respectively (scale bars, 25 μm). Bars represent means ± SD; unpaired Student’s t-test, ***p<0.001.
-
Figure 8—source data 1
Original blot images of Figure 8.
- https://cdn.elifesciences.org/articles/86689/elife-86689-fig8-data1-v1.zip
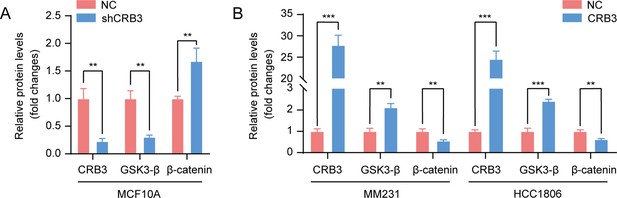
Western blotting quantification of the molecules in the Wnt signaling pathway.
(A) The relative protein levels of CRB3, GSK3-β, and β-catenin in CRB3-depleted MFC10A cells (normalized to β-actin). (B) The relative protein levels of CRB3, GSK3-β, and β-catenin in MDA-MB-231 and HCC 1806 breast cancer cells with CRB3b overexpression (normalized to β-actin). The data are three independent experiments performed in triplicate. Bars represent means ± SD; unpaired Student’s t-test, **p<0.01, ***p<0.001.

Schematic model of CRB3 regulating ciliary assembly.
Graphic summary of prominent phenotypes observed after CRB3 deletion. CRB3 is localized on apical epithelial surfaces and participates in tight junction formation to maintain contact inhibition and cell homeostasis in quiescence. Inside the cell, Rab11-positive endosomes mediate the intracellular trafficking of CRB3, and CRB3 navigates GCP6/Rab11 trafficking vesicles to CEP290. Then, GCP6 is involved in normal γ-tubulin ring complex (γTuRC) assembly in ciliogenesis. In CRB3 deletion cells, the primary cilium fails to assemble properly, and the Wnt signaling pathway is activated through β-catenin upregulation and nuclear localization, but the Hh signaling pathway fails to be activated. This cellular imbalance is disrupted, leading to tumorigenesis. TJ, tight junction; EE, early endosome; LE, late endosome.
Additional files
-
Supplementary file 1
The sequences of primer pairs used in real-time PCR.
- https://cdn.elifesciences.org/articles/86689/elife-86689-supp1-v1.docx
-
Supplementary file 2
Characteristics of patients.
- https://cdn.elifesciences.org/articles/86689/elife-86689-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/86689/elife-86689-mdarchecklist1-v1.docx





