DnaJC7 specifically regulates tau seeding
Figures
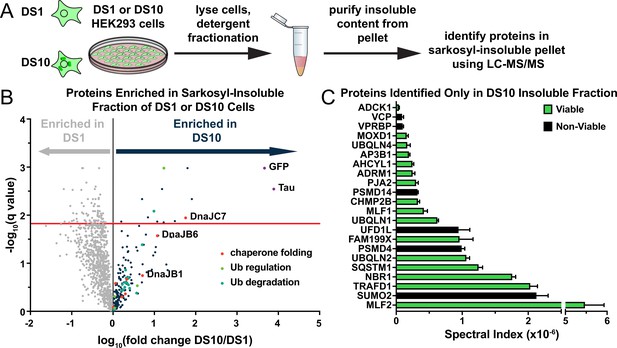
A proteomic approach to identify tau aggregate interactors.
(A) Tau aggregates were partially purified from DS1 and DS10 HEK293 cells expressing tauRD-YFP. Detergent fractionation enabled generation of a sarkosyl-insoluble fraction containing tau aggregates. Proteins were resolved by SDS-PAGE and then extracted from individual lanes for analysis by LC-MS/MS. (B) Volcano plot showing proteins enriched in the sarkosyl-insoluble fraction as a fold enrichment from cells expressing tauRD-YFP aggregates (DS10, dark blue dots) over cells expressing tauRD-YFP that does not form aggregates (DS1, gray dots). The red line indicates a false discovery rate of 1.5%. Gene ontology (GO) term enrichment analyses of biological processes is also shown for select GO terms: orange dots, chaperone-mediated protein folding (chaperone folding); green dots, regulation of ubiquitination (Ub regulation); teal dots, ubiquitin-dependent protein catabolic process (Ub degradation). (C) Spectral indices for a selection of the proteins identified only in the DS10-insoluble fraction. Viable knockouts are shown as green bars. Non-viable knockouts are shown as black bars. Error bars represent the SEM of three extracted protein SDS-PAGE gel bands.
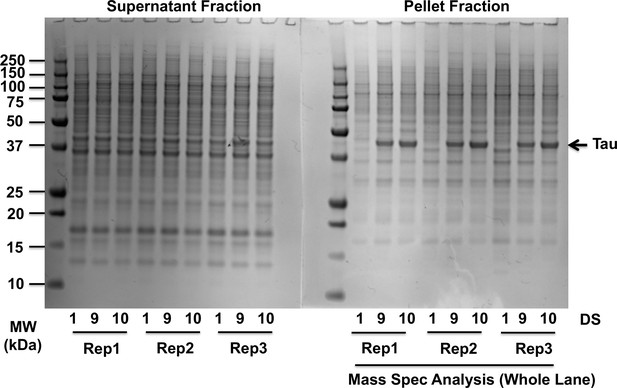
Partial purification of tau aggregates.
Sarkosyl-soluble and sarkosyl-insoluble fractions were run on gels and proteins were stained with SimplyBlue protein stain. DS9 and DS10 featured significant enrichment of insoluble tauRD-YFP (arrow on gel). Whole lanes for pellet fractions were analyzed by mass spectrometry (biological triplicates). The DS9 cell line, which contains tau aggregates of a distinct strain, is included as a tau-aggregate containing control. Source data for this figure are provided in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
This source data file contains the original uncropped images for the SDS-PAGE gels shown in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/86936/elife-86936-fig1-figsupp1-data1-v2.zip
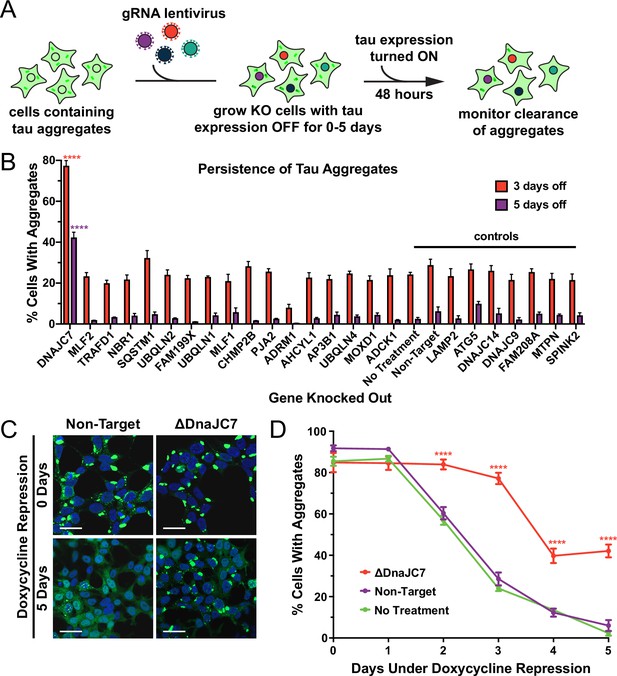
DnaJC7 knockout (KO) uniquely extends tau seed lifespan in dividing cells.
(A) Schematic showing the HEK293 OFF1::DS10 system. A selection of the hits from the proteomics screen was knocked out in these cells. The cells are then allowed to grow with tau expression turned OFF for 0–5days before resuming tau expression. Error bars represent the SEM of six technical replicates. (B) The persistence of tau aggregates in OFF1::DS10 cells with the indicated KO was quantified following 3 (orange bars) or 5 (purple bars) days of repressed tau expression. Error bars represent the SEM of six technical replicates. (C) Confocal microscopy images showing tau aggregate organization in the DnaJC7 KO and nontargeting control cells following 0 or 5days of repression of tau expression. Scale bars denote 20 µm. (D) Extended time course for tau aggregate clearance in the OFF1::DS10 system with DnaJC7 KO (orange) and the nontargeting (purple) and untreated (green) controls. Error bars represent SEM of six technical replicates. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
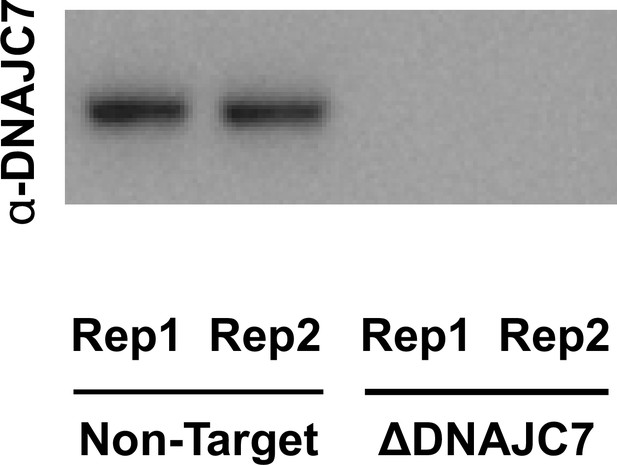
DnaJC7 knockout (KO) in (OFF1::DS10) cells.
Immunoblotting for DnaJC7 confirms that DnaJC7 is knocked out in the Tet-regulated tauRD-YFP aggregate line (OFF1::DS10). Source data for this figure are provided in Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
This source data file contains the original uncropped images for the western blot shown in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/86936/elife-86936-fig2-figsupp1-data1-v2.zip
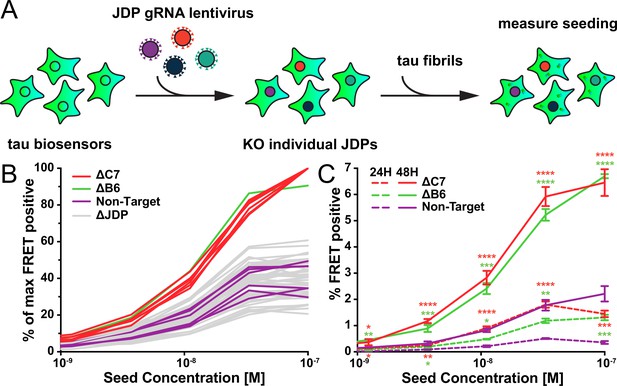
A targeted genetic screen for modifiers of transient tau seeding identifies specific J domain proteins (JDPs).
(A) Schematic showing the HEK293T tau biosensor system consisting of tauRD fused to either mCerulean3 or mClover3 fluorescent proteins. The biosensor cells had each JDP individually knocked out to generate 50 distinct cell lines. Recombinant, sonicated tau fibrils (seeds) were added to the cells to induce seeding of the tauRD constructs, which was detected as a FRET signal via flow cytometry 48hr after treatment. (B) Representative data showing the effects of the individual knockouts (KO) of JDPs on tau seeding in biosensor cells, quantified via flow cytometry. Cells were seeded with a dose titration of sonicated tau fibrils. KO of DnaJC7 (ΔC7, orange) and DnaJB6 (ΔB6, green) are highlighted. The remaining JDP KO are denoted as ΔJDP (gray). Each batch of KO cell lines was normalized to the DnaJC7 KO seeding signal and then all batches are plotted together. The seeding assay for the DnaJC7 KO and the nontargeting control (Non-Target, purple) were repeated 10 times. (C) Extended time course harvesting of the tau seeding assay for DnaJB6 KO, DnaJC7 KO, and nontargeting control cells harvested at 24hr (24H, dashed lines) and 48hr (48H, solid lines) timepoints. Coloring as in (B). Error bars represent SEM of three technical replicates. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
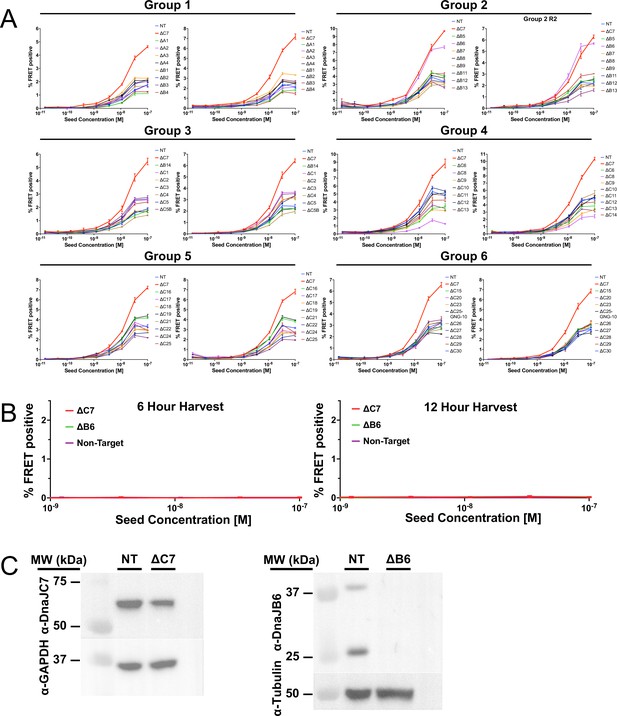
All individual groups of the J domain protein (JDP) CRISPR screen.
(A) Full tau dose titrations for all batches of individual knockouts (KO) of JDPs on tau seeding in biosensor cells, quantified by FRET signal via flow cytometry. Cells were seeded with a dose titration of sonicated tau fibrils. KO of DnaJC7 (ΔC7, red) and the nontargeting control (NT, blue) are highlighted in each batch. (B) Extended time course harvesting of the tau seeding assay for DnaJC7 KO (ΔC7, orange), DnaJB6 KO (ΔB6, green), and nontargeting control (Non-Target, purple) cells at 6hr and 12hr timepoints. All error bars represent SEM of three technical replicates. (C) Immunoblotting for DnaJC7 and DnaJB6 confirms partial and full KO in their respective tau biosensor cell lines, ΔB6 and ΔC7. The nontargeting control cell line (NT) is also shown as a control. Source data for this figure are provided in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
This source data file contains the original uncropped images for the western blots shown in Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/86936/elife-86936-fig3-figsupp1-data1-v2.zip
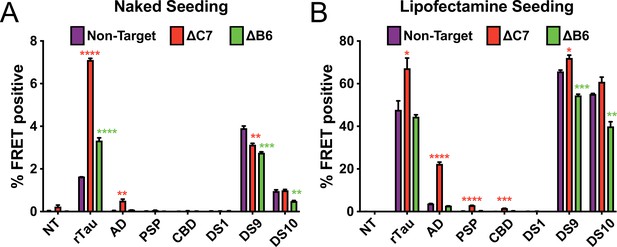
DnaJC7 knockout (KO) increases tau seeding across multiple seed sources.
Intracellular (A) naked or (B) lipofectamine-mediated tau seeding in nontargeting control (Non-Target, purple), DnaJC7 KO (ΔC7, orange), and DnaJB6 KO (ΔB6, green) tau biosensor cells. Cells were seeded with sonicated recombinant tau fibrils (rTau); brain lysates from patients with Alzheimer’s disease (AD), progressive supranuclear palsy (PSP), or corticobasal degeneration (CBD); and cell lysates of DS1, DS9, or DS10 cells. 100nM or 10nM of recombinant tau were added for naked and Lipofectamine seeding, respectively. 25μL or 5μL of patient brain or lysate or 20μg or 5μg cell lysate were added for naked and Lipofectamine seeding, respectively. NT denotes no treatment with seeds. Error bars represent SEM of three technical replicates. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
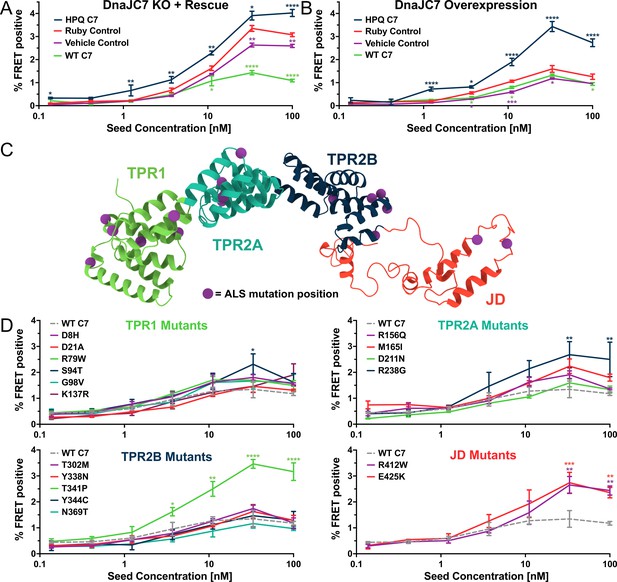
DnaJC7 regulates tau seeding in multiple experimental approaches.
(A) Rescue of DnaJC7 knockout (KO) in tau biosensor cells with either wildtype (WT C7, green) or HPQ mutant (HPQ C7, dark blue) DnaJC7 constructs. The Ruby fluorophore alone (Ruby Control, orange) and a vehicle control (Vehicle Control, purple) were also added to the DnaJC7 KO cells. (B) Overexpression of DnaJC7 constructs in control tau biosensor cells. The cells were transfected with the same constructs as in (A). (C) Model of DnaJC7 with domains colored as follows: TPR1, green; TPR2A, teal; TPR2B, dark blue; JD, orange. Positions of amyotrophic lateral sclerosis (ALS)-associated mutations are shown as purple spheres. (D) Rescue of DnaJC7 KO in tau biosensor cells with ALS-associated mutants of DnaJC7 and WT control, sorted by domain location. Rescue with the WT DnaJC7 construct is shown in all domains as a gray, dashed line. Error bars represent SEM of three technical replicates. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
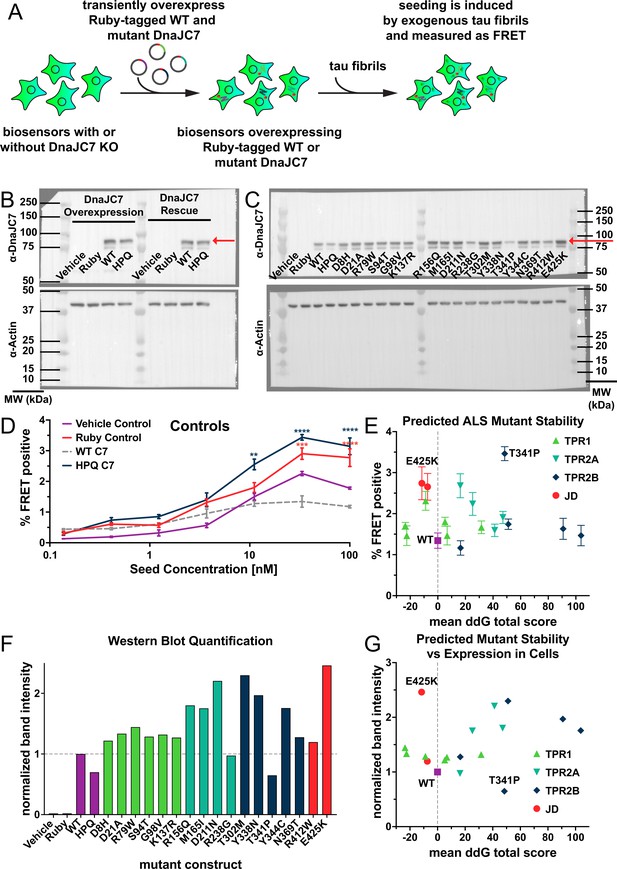
Stability and expression of DnaJC7 mutants in tau biosensors.
(A) Schematic showing tau biosensor cells with or without DnaJC7 knockout (KO) transiently overexpressing different Ruby-tagged gRNA-resistant DnaJC7 constructs. Cells were allowed to express the constructs for 2 days before being plated for the seeding assay. (B) Immunoblotting for DnaJC7 confirmed expression of the Ruby-WT DnaJC7 and Ruby-DnaJC7 (HPQ) mutant constructs in tau biosensor cells without (Overexpression) and with endogenous DnaJC7 knocked out (Rescue). Ruby fusion constructs are highlighted by a red arrow. (C) Immunoblotting for DnaJC7 confirmed expression of the Ruby-WT DnaJC7 and Ruby-DnaJC7 amyotrophic lateral sclerosis (ALS) mutant constructs in tau biosensor cells with endogenous DnaJC7 knocked out. Ruby fusion constructs are highlighted by a red arrow. (D) Positive and negative controls utilized in the rescue of DnaJC7 KO in tau biosensor cells with ALS-associated mutants of DnaJC7, colored as follows: Vehicle Control, purple; Ruby Control, orange; WT DnaJC7, gray dashed; HPQ mutant, dark blue. Error bars represent SEM of three technical replicates. (E) Rosetta-calculated mean Gibbs free energy shift (ddG) of the ALS-associated mutants of DnaJC7 vs. their rescue seeding with 33nM of tau fibrils. Gray dashed line denotes a mean ddG total score of 0. Mutants are colored according to their domain localization: TPR1, green; TPR2A, teal; TPR2B, dark blue; JD, orange. (F) Quantification of the western blot signal for the different ALS-associated mutants and controls in (C). All constructs were normalized to the band intensity of the WT construct. Domains are colored as in (C). (G) Rosetta-calculated mean Gibbs free energy shift (ddG) of the ALS-associated mutants of DnaJC7 vs. their normalized western blot band intensity. Gray dashed line denotes a mean ddG total score of 0. Mutants are colored as in (C).All error bars represent SEM of three technical seeding replicates. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001. Source data for this figure are provided in Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
This source data file contains the original uncropped images for the western blots shown in Figure 5—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/86936/elife-86936-fig5-figsupp1-data1-v2.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | HEK293 | ATCC | CRL-1573 | |
| Cell line (human) | HEK293T | ATCC | CRL-3216 | |
| Cell line (human) | HEK293T tauRD P301S v2L FRET Biosensor | Produced by Diamond Lab | Stably expresses mClover3 and mCerulean3 tagged tauRD monomers. | |
| Cell line (human) | HEK293 DS1 tauRD(P301L-V337M)-YFP | Produced by Diamond Lab | Stably propagates diffuse tauRD monomers. | |
| Cell line (human) | HEK293 DS10 tauRD(P301L-V337M)-YFP | Produced by Diamond Lab | Stably propagates a unique tau strain. | |
| Cell line (human) | HEK293 OFF1::DS10 tauRD(P301L-V337M)-YFP | Produced by Diamond Lab | Stably propagates a unique tau strain under a tetracycline-repressible promoter. | |
| Biological sample (human) | Alzheimer’s Disease subject brain | UT Southwestern Alzheimer’s Disease Center | ||
| Biological sample (human) | Progressive Supranuclear Palsy subject brain | UT Southwestern Alzheimer’s Disease Center | ||
| Biological sample (human) | Corticobasal Degeneration subject brain | UT Southwestern Alzheimer’s Disease Center | ||
| Antibody | Rabbit polyclonal anti-DnaJC7 | Proteintech | 11090-1-AP | 1:2000 dilution |
| Antibody | Rabbit polyclonal anti-DnaJB6 | Proteintech | 11707-1-AP | 1:2000 dilution |
| Antibody | Rabbit polyclonal anti-Beta-Tubulin | Proteintech | 10094-1-AP | 1:5000 dilution |
| Antibody | Donkey-anti-rabbit HRP- linked F(ab’)2 | Cytiva | NA9340-1ML | 1:5000 dilution (DnaJC7, Beta-Tubulin), 1:4000 dilution (DnaJB6) |
| Antibody | Mouse monoclonal anti-GAPDH | Proteintech | 60004-1-Ig | 1:10000 |
| Antibody | Mouse monoclonal anti-Beta-Actin | Proteintech | 66009-1-Ig | 1:5000 dilution |
| Antibody | Goat-anti-mouse H&L (HRP) | Abcam | ab6789 | 1:10000 dilution (GAPDH), 1:5000 dilution (Beta-Actin) |
| Peptide, recombinant protein | Human tau 2N4R (Full Length WT-tau) fibrils | Produced by Diamond Lab | MAEPRQEFEVMEDHAGTYGLGDRKDQGGYTMHQDQEGDTDAGLKESPLQ TPTEDGSEEPGSETSDAKSTPTAEDVTAPLVDEGAPGKQAAAQPHTEIPEGTT AEEAGIGDTPSLEDEAAGHVTQARMVSKSKDGTGSDDKKAKGADGKTKIATP RGAAPPGQKGQANATRIPAKTPPAPKTPPSSGEPPKSGDRSGYSSPGSPGTP GSRSRTPSLPTPPTREPKKVAVVRTPPKSPSSAKSRL | |
| Recombinant DNA reagent | gRNA-resistant WT DnaJC7 cDNA sequence | gBlock from IDT, cloned by VAP | The sequence can be found in Materials and methods: Design of a DnaJC7 construct resistant to targeting by used gRNA sequences |





