Control of telomere length in yeast by SUMOylated PCNA and the Elg1 PCNA unloader
Figures
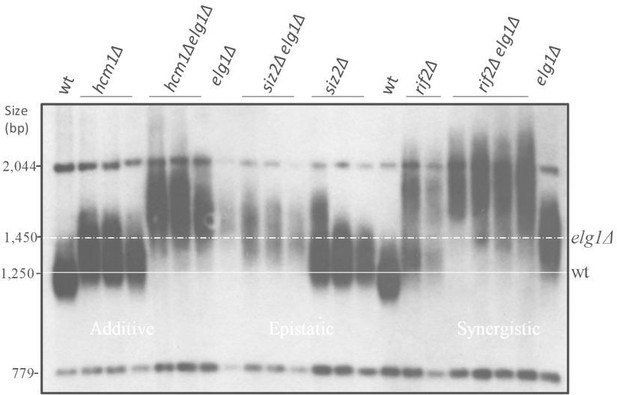
SUMOylated PCNA regulates telomere length.
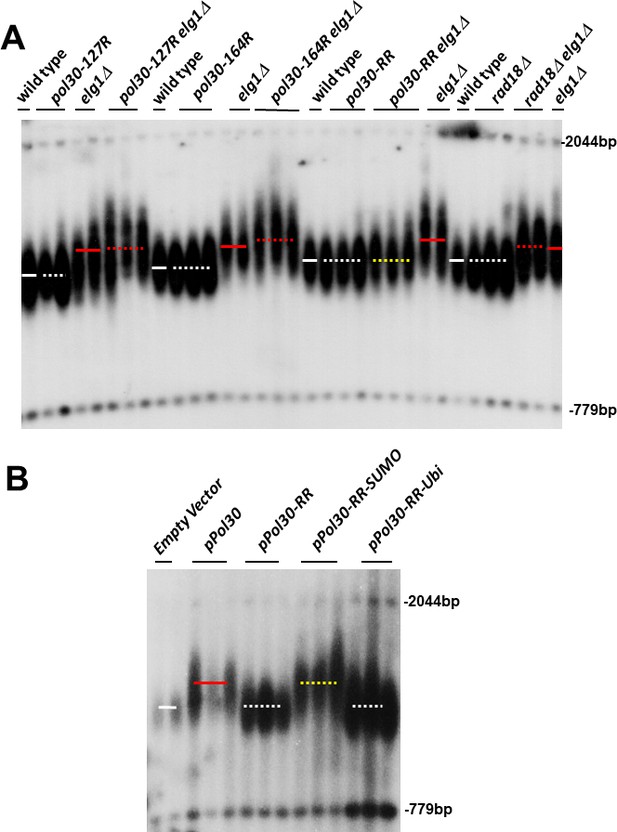
(A) Southern blot (Teloblot) showing that lack of SUMOylation of PCNA prevents telomere elongation. Independently created colonies were passaged ten times, its DNA extracted, digested with XhoI, and run in an agarose gel. The DNA was then transferred to a nitrocellulose membrane, which was incubated with a radioactive probe that detects telomeres, and a size marker. (B) Teloblot showing that overexpression of wild-type PCNA or Pol30-RR-SUMO fusion, but not Pol30-ubiquitin fusion or Pol30-RR causes telomere elongation.
-
Figure 1—source data 1
Source data is the original Southern blot.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig1-data1-v1.zip
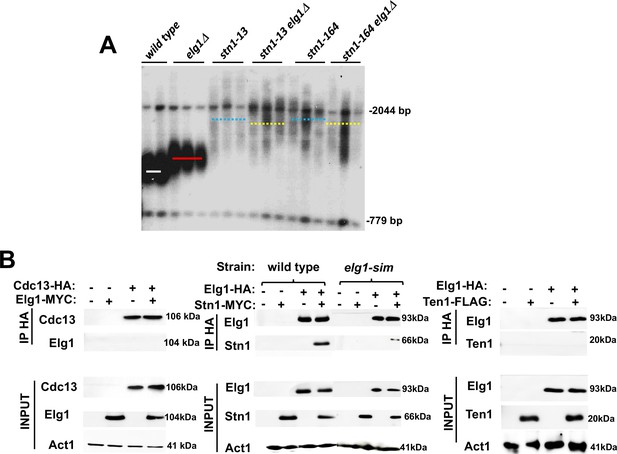
Genetic and physical interaction between ELG1 and STN1.
(A) Teloblot showing epistasis between elg1Δ and stn1 mutants. (B) Co-Immunoprecipitation experiment showing physical interaction between Elg1 and Stn1 and reduced physical interaction between Elg1-sim and Stn1. No interaction could be detected between Elg1 and Cdc13 or Ten1.
-
Figure 2—source data 1
The source data contains the Southern blots and the Western blots used to make the figure.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig2-data1-v1.zip
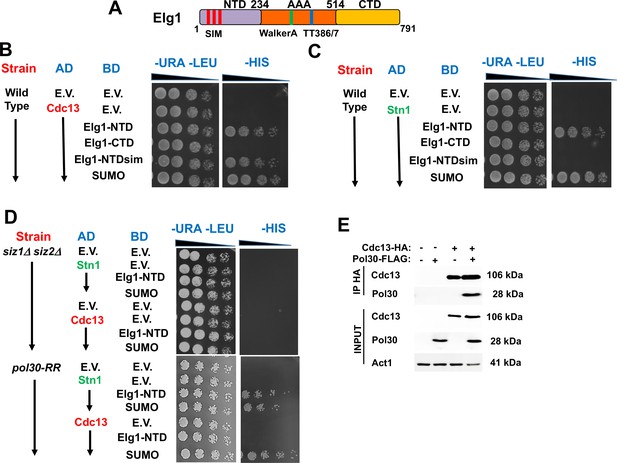
Determinants of the Elg1-Stn1 interaction.
(A) Schematic representation of the Elg1 protein. The three SIM motifs, the WalkerA motif and the two threonines at the interface with PCNA, are shown. (B) Yeast two-hybrid (YTH) interaction of Cdc13 and Elg1 in a wild-type strain. AD: protein fused to the activating domain of Gal4; BD: protein fused to the DNA binding domain of Gal4. e.v.: empty vector. (C) YTH interaction of Stn1 and Elg1 in a wild-type strain. (D) YTH experiments in the siz1Δ siz2Δ and pol30-RR background. (E) Co-Immunoprecipitation experiment showing physical interaction between Cdc13 and PCNA.
-
Figure 3—source data 1
The Source data contains the Western blots used in Figure 3E.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig3-data1-v1.zip

Weak interaction between Elg1 and Ten1.
Results of yeast two-hybrid (YTH) experiments.

Cdc13 interacts with PCNA, and mutations that prevent PCNA SUMOylation (pol30-RR) also impair its interaction with Cdc13.

The unloading activity of Elg1 and Cdc13 SUMOylation are necessary for the Elg1-Cdc13 interaction.
(A) Yeast-two-hybrid (YTH) experiment in a elg1Δ strain. (B) YTH experiment in a elg1-sim strain. (C) YTH experiment in a elg1-TT386/7DD strain. (D) YTH experiment in a elg1-sim+DD strain. (E) YTH experiment in a elg1-Walker AB strain. (F) Lack of interaction between cdc13-snm and Elg1 (in a wild-type strain).
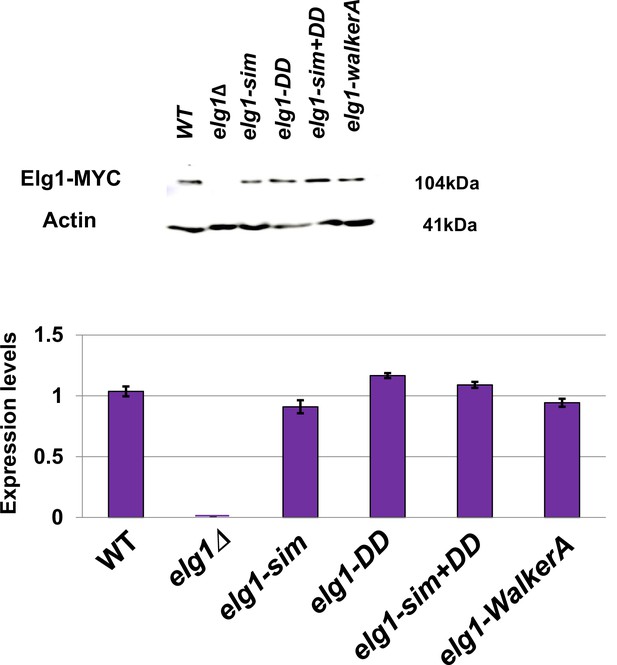
All elg1 alleles used are expressed at similar levels.
Western blot results.
-
Figure 4—figure supplement 1—source data 1
The Source Data contains the Western Blot used in the figure.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig4-figsupp1-data1-v1.zip
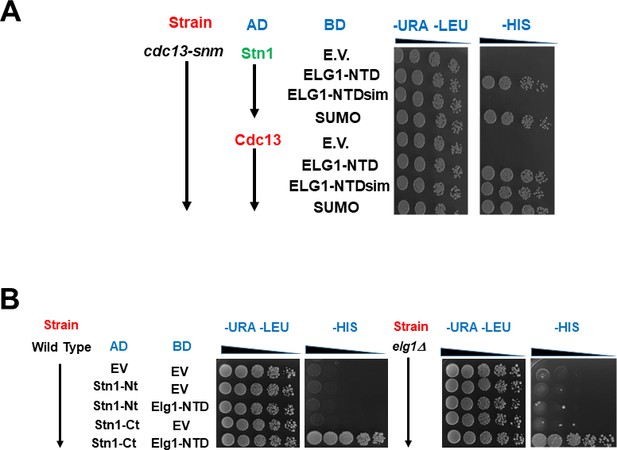
SUMOylation of the genomic copy of Cdc13 has no effect on the interactions of a wt copy with Stn1 or Elg1.
(A) The sumo-no-more allele of Cdc13 has no effect on the yeast-two-hybrid (YTH) interactions between wt Cdc13 or Stn1 and the N-terminus of Elg1. (B) The C-terminus of Stn1 interacts with the N-terminus of Elg1 in wild-type and elg1Δ strains.
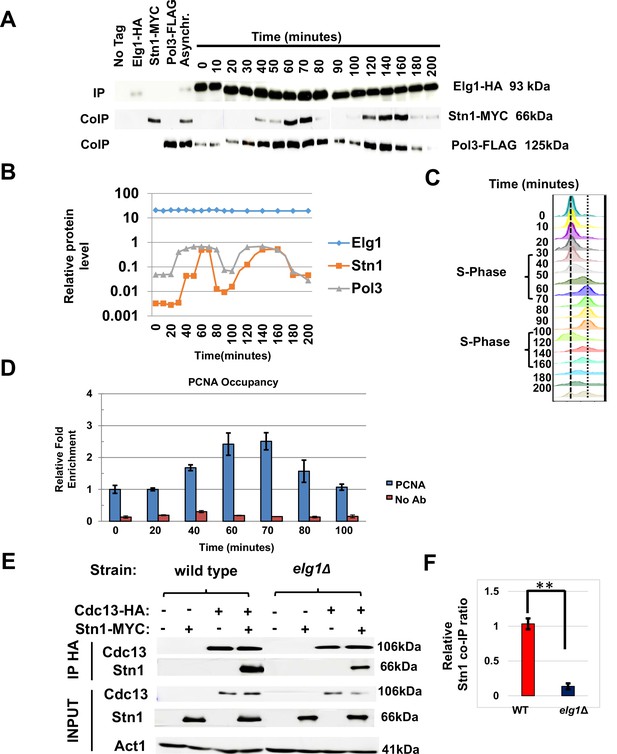
Timing of Elg1-Stn1 interaction.
(A) Co-IP experiment with synchronized cells. Aliquots were taken at time intervals, Elg1 was immunoprecipitated, and the level of Stn1 and Pol3 (the large subunit of DNA polymerase Delta) was monitored by western blot. Strains with single tags are shown as controls. Whole-cell extract results are shown in Figure 5—figure supplement 1. (B) Quantitation of the western shown in (A). (C) DNA content of the cells used in (A) by cell cytometry. (D) Chromatin immunoprecipitation at telomeres (Telo-ChIP) in synchronized cells showing PCNA occupancy. (E) Interaction between Stn1 and Cdc13 in a wild type and a elg1Δ strain. (F) Quantitation of three independent biological repeats of the experiment shown in (E). **p<0.001.
-
Figure 5—source data 1
The Source Data contains the original Western blots used to make the figure.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig5-data1-v1.zip
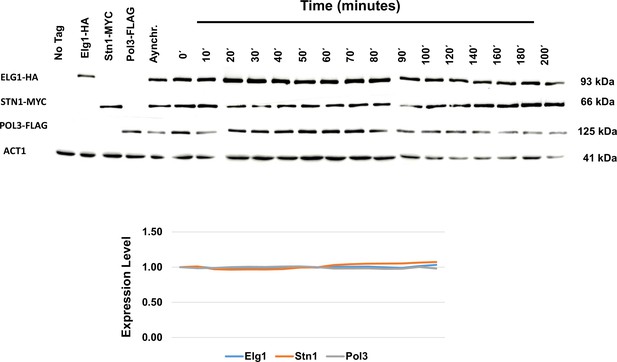
Whole-cell extract showing the level of Elg1, Stn1, and Pol3 in the cell cycle experiment shown in Figure 5.
-
Figure 5—figure supplement 1—source data 1
The Source Data contains the original Western blots used to generate the figure.
- https://cdn.elifesciences.org/articles/86990/elife-86990-fig5-figsupp1-data1-v1.zip
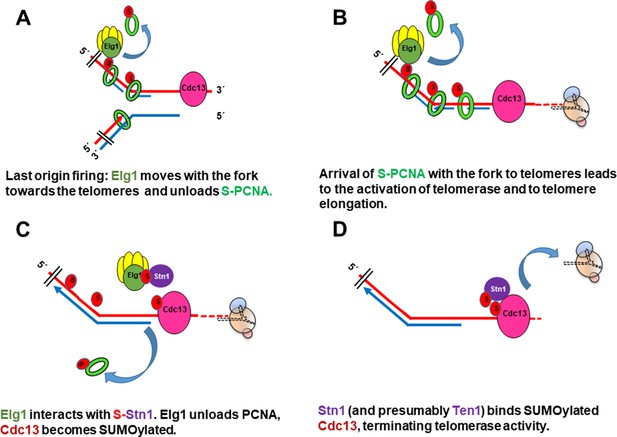
Model for the role of SUMOylated PCNA and Elg1 in telomere length regulation.
(A) Cdc13 binds ssDNA at the telomeres, Elg1 moves with the replisome at the lagging strand, unloading PCNA in each Okazaki fragment. (B) Arrival of the SUMOylated PCNA at the fork to Cdc13 promotes telomerase activity. (C) Elg1 interacts with Stn1, which could be SUMOylated. Cdc13 becomes SUMOylated. Elg1 unloads PCNA and leaves telomeres. (D) Stn1 is now able to interact with Cdc13, evicting Est1 and terminating telomerase activity.
Tables
Summary of all yeast two-hybrid (YTH) interactions presented.
| Strain | Plasmid | Interaction w/Stn1 | Interaction w/Cdc13 |
|---|---|---|---|
| Wild type | Elg1-NTD | Yes | Yes |
| Wild type | Elg1-sim | No | Yes |
| siz1Δ siz2Δ | Elg1-NTD | No | No |
| pol30-RR | Elg1-NTD | Yes | No |
| elg1Δ | Elg1-NTD | Yes | No |
| elg1-sim | Elg1-NTD | Yes | Yes |
| elg1-DD | Elg1-NTD | Yes | No |
| elg1-sim+DD | Elg1-NTD | Yes | No |
| elg1-WalkerA | Elg1-NTD | Yes | No |
| cdc13-snm | Elg1-NTD | Yes | Yes |
| Wild type | Elg1-NTD | - | snm: no |
-
elg1-sim: mutation in the SUMO-interacting motif of Elg1; elg1-DD: TT386/7DD; elg1-sim+DD: combination of mutations in the SIM and in TT386/7; elg1-WalkerA: mutation that eliminates ATPase and unloading activity of Elg1. cdc13-snm: allele of Cdc13 that cannot be SUMOylated.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pGBD424 | Takahashi et al., 2020 | Empty yeast two-hybrid vector | |
| Recombinant DNA reagent | pGBD424-Pol30 | Takahashi et al., 2020 | Overexpresses wt PCNA | |
| Recombinant DNA reagent | pGBD424-Pol30-RR | Takahashi et al., 2020 | Overexpresses unmodifiable PCNA | |
| Recombinant DNA reagent | pGBD424-ubiquitin-Pol30-RR | Takahashi et al., 2020 | Overexpresses unmodifiable PCNA fused to ubiquitin | |
| Recombinant DNA reagent | pGBD424-SUMO-Pol30-RR | Takahashi et al., 2020 | Overexpresses unmodifiable PCNA fused to SUMO | |
| Recombinant DNA reagent | pGBU9 | Parnas et al., 2010 | Yeast two-hybrid vector | |
| Strain, strain background (Saccharomyces cerevisiae MATa strain) | PJ69-4A | James et al., 1996 | Yeast two-hybrid strain | |
| Chemical compound, drug | Alpha Factor | Sigma-Aldrich | T6901 | |
| Commercial assay or kit | Protein A sepharose beads | Sigma-Aldrich | 17-1279-01 | |
| Commercial assay or kit | Protein G sepharose beads | Sigma-Aldrich | 17-0618-01 | |
| Chemical compound, drug | Pronase | Sigma-Aldrich | P5147 | |
| Antibody | Anti-HA (mouse polyclonal) | Santa Cruz Biotechnology | sc7392 | 1:1000 |
| Antibody | Anti-Myc (mouse polyclonal) | Santa Cruz Biotechnology | 9E10, SC-40 | 1:1000 |
Yeast strains used in this study.
| Strain number | Name | Genotype |
|---|---|---|
| 13237 | BY4741 | MATa his3del, leu2del, met15del, ura3del |
| 12480 | BY elg1Δ::HygMX | MATa his3del, leu2del, met15del, ura3del, elg1::HYG |
| 14421 | BY pol30-K127R | MATa his3del, leu2del, met15del, ura3del, pol30-K127R::LEU2 |
| 14425 | BY pol30-K164R | MATa his3del, leu2del, met15del, ura3del, pol30-K164R::LEU2 |
| 14423 | BY pol30-KK127,164RR | MATa his3del, leu2del, met15del, ura3del, pol30-RR::LEU2 |
| 14426 | BY pol30-K127R elg1Δ | MATa his3del, leu2del, met15del, ura3del, pol30-K127R::LEU2, elg1::HYG |
| 14430 | BY pol30-K164R elg1Δ | MATa his3del, leu2del, met15del, ura3del, pol30-K164R::LEU2, elg1::HYG |
| 14428 | BY pol30-KK127,164RR elg1Δ | MATa his3del, leu2del, met15del, ura3del,pol30-RR::LEU2,elg1::HYG |
| 14398 | BY rad18Δ:: KanMX | MATa his3del, leu2del, met15del, ura3del, rad18::KanMX. |
| 14401 | BY rad18D::KanMX elg1D::HygMX | MATa his3del, leu2del, met15del, ura3del, rad18::KanMX, elg1::HYG |
| 19606 | BY pol30-KK127,164RR Elg1-HA | MATa his3del, leu2del, met15del, ura3del ELG1-HA-NAT pol30-KK127,164RR:HIS3 |
| 20622 | BY bar1 CDC13-HA ELG1-HA | MATa his3del, leu2del, met15del, ura3del, bar1::LEU2, CDC13:3HA:HISMX, ELG1-HA-KanMX |
| 18418 | BY STN1-Myc Elg1-HA | MATa his3del, leu2del, met15del, ura3del stn1::HYG CEN LEU2 STN1-(G)9-(myc)7 ELG1-HA-KanMx |
| 18790 | BY bar1 STN1-Myc ELG1-HA POL3-FLAG | MATa his3del, leu2del, met15del, ura3del bar1::NatMX stn1::HYG CEN LEU2 STN1-(G)9-(myc)7 ELG1-HA-KanMx POL3-FLAG-URA3 |
| 19552 | BY bar1 CDC13-HA POL30-FLAG | MATa his3del, leu2del, met15del ura3del, bar1:: NatMX CDC13-3HA::HISMX, POL30-FLAG::KanMX, |
| 20625 | BY CDC13-HA STN1-Myc | MATa his3del, leu2del, met15del, ura3del, CDC13-3HA::HISMX, stn1::HYG CEN LEU2 STN1-(G)9-(myc)7, |
| 20626 | BY CDC13-HA STN1-Myc elg1D | MATa his3del, leu2del, met15del, ura3del,CDC13-3HA::HISMX, stn1::HYG,CEN LEU2 STN1-(G)9-(myc)7,elg1::KanMx |
| 20623 | BY TEN1-FLAG | MATa his3del, leu2del, met15del, ura3del trp1del Lys2del can1:: STE2pr-Sp_HIS5 ten1::KanMX, Elg1-HA-NAT CEN URA3 TEN1-(G)8-(FLAG)3 |
| 17611 | W303-1a | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100 |
| 9842 | W303 elg1Δ::HygMX | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100, elg1::KanMx |
| 9551 | W303 stn1-13 | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100, stn1-13 |
| 9848 | W303 elg1Δ::HygMX stn1-13 | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100 elg1::KanMx, stn1-13 |
| 12357 | W303 stn1-164 | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100, stn1Δ::Hyg+pRS-stn1-L164A (pVL3571) |
| 12358 | W303 stn1-164 elg1Δ: KanMX | MATa leu2-3, 112 ura3-1 his3-11,15, trp1-1, ade2-1, can1- 100, elg1::KanMx, stn1Δ::Hyg+pRS-stn1-L164A (pVL3571) |
| 12062 | PJ69-4 | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ |
| 15017 | PJ elg1D | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ elg1::HYG |
| 19774 | PJ siz1 siz2 | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ siz1::KanMX siz2::HYG |
| 11069 | PJ pol30-RR | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ POL30-RR:: leu2:: KANMX |
| 20624 | PJ STN1-Myc | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ elg1-sim::KanMx, stn1::HYG,CENLEU2 STN1-(G)9-(myc)7 |
| 19916 | PJ elg1-sim-Myc | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ elg1-SIM-MYC-KANMX |
| 18798 | PJ elg1-DDMyc | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ elg1-DD-MYC-KANMX |
| 19917 | PJ elg1-DD+sim-Myc | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ elg1-DDsim-MYC-KANMX |
| 19915 | PJ elg1-Walker A | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ, elg1 KK343/4DD:MYC:KanMX |
| 19486 | PJ cdc13-snm | MAT@ trp1-901 leu2-3,112 ura3-52 his3-200 gal4del gal80del GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ cdc13-snm |
Plasmids used in this study.
| Strain number | Genotype | Source or reference |
|---|---|---|
| 4239 | pGad424 (LEU2) | Kupiec lab |
| 4238 | pGad424-POL30 | Helle Ulrich lab |
| 4237 | pGad424-pol30-k127r,k164r | Helle Ulrich lab |
| 4236 | pGad424-pol30-k127r,k164r-SMT3 | Helle Ulrich lab |
| 4235 | pGAD424-pol30-k127r,k164r-UBI | Helle Ulrich lab |
| 4147 | CEN LEU2 STN1-(G)9-(myc)7 | Victoria Lundblad lab |
| 4144 | CEN URA3 TEN1-(G)8-(FLAG)3 | Victoria Lundblad lab |
| 2201 | pCN181 pACT2-STN1 (LEU2) | Constance Nugent lab |
| 2205 | pVL855 pACT2-CDC13 | Constance Nugent lab |
| 2168 | pACT2 - TEN1 | Michel Charbonneau lab |
| 1775 | pGBU9 (URA3) | Kupiec lab |
| 1973 | pGBU9-ELG1-NTD(1-234) | This study |
| 3301 | pGBU9-ELG1-CTD(541-791) | This study |
| 2260 | pGBU9-ELG1-NTDsim(1-234) | This study |
| 2241 | pGBU9-SMT3 | This study |
| 2419 | pGAD424-Stn1-Nt(1-282) | David Shore lab |
| 2420 | pGAD424-Stn1-Ct(282-494) | David Shore lab |
| 2169 | pGAD424-Stn1-13 | Michel Charbonneau lab |
| 2418 | pGAD424-Stn1-63 | David Shore lab |
| 4287 | pACT-cdc13-snm | This study |