Anti-inflammatory therapy with nebulized dornase alfa for severe COVID-19 pneumonia: a randomized unblinded trial
Figures
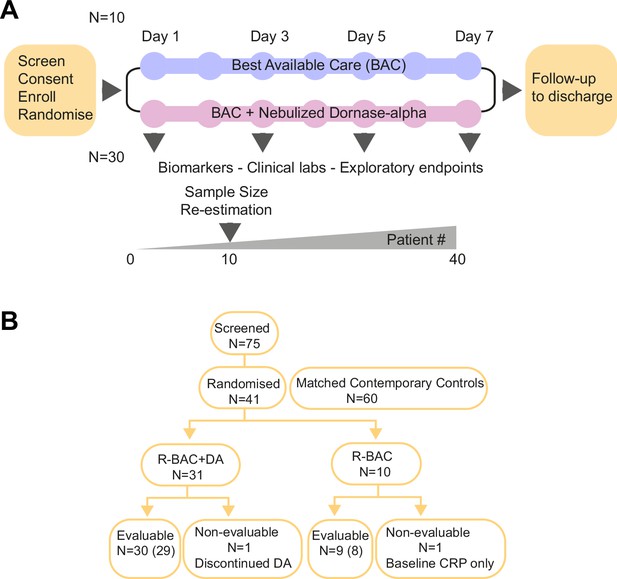
Trial design and Consort diagram.
(A) COVASE Trial Design. (B) Consort diagram summary. Numbers not in parentheses indicate the participants in the intention-to-treat (ITT) population and the numbers in parentheses indicate the number of participants in the per-protocol population. A complete consort flow diagram is shown in Figure 1—figure supplement 1.
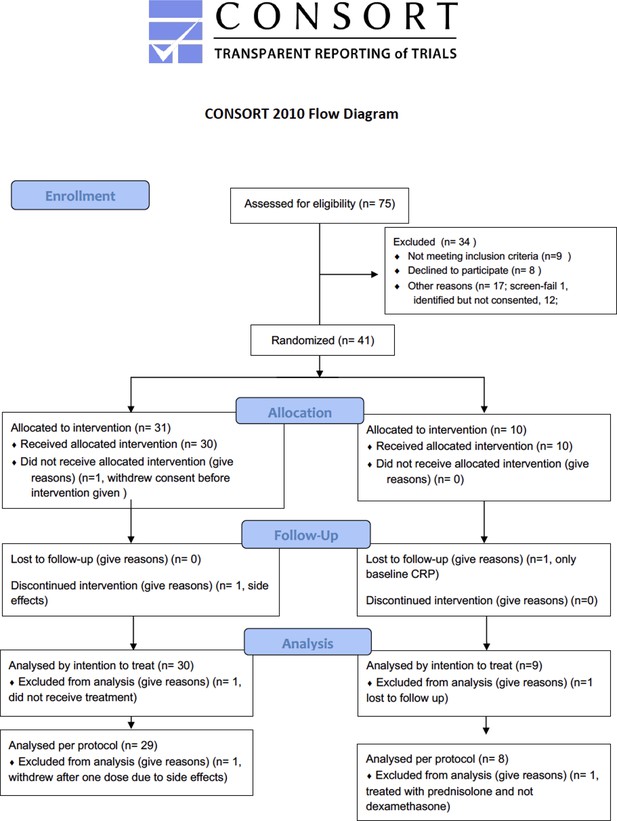
Consort flow diagram.
Flow diagram of recruited randomized participants in the randomised to BAC (R-BAC) and R-BAC +DA arms, depicting the allocation following randomization, the numbers included and lost to follow-up and the numbers of included and excluded participants by intention to treat and per protocol and the justification for each exclusion.
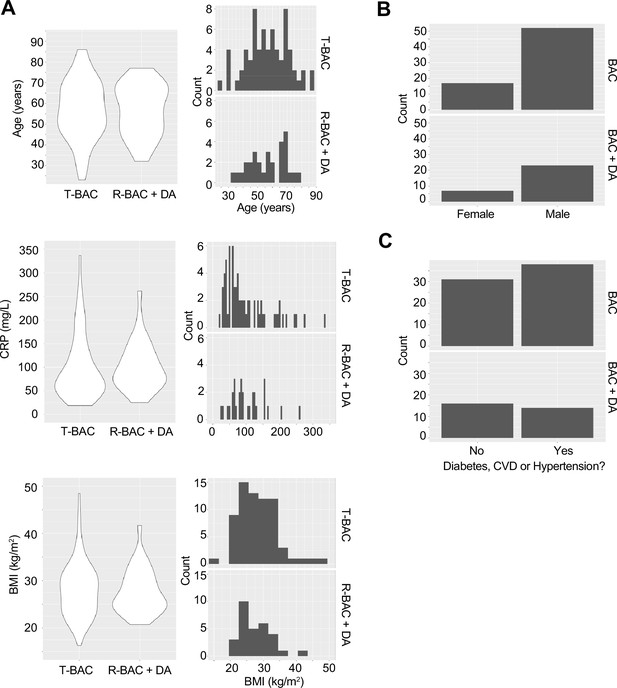
Baseline characteristics of patients analysed in the trial.
(A) Violin plots (left) and frequency distribution (right) of baseline clinical parameters between participants in the contemporary control and randomized best available care (BAC) group (T-BAC) and the randomized BAC +Dornase alfa (R-BAC +DA) group. (Top) Age, (middle) Baseline C-reactive protein (CRP) and (bottom) Body mass index (BMI). (B) Number of male and female participants in the two groups. (C) Incidence of cardiovascular comorbidities in the two groups.
-
Figure 1—figure supplement 2—source data 1
Baseline characteristics in T-BAC and R-BAC +DA participants.
- https://cdn.elifesciences.org/articles/87030/elife-87030-fig1-figsupp2-data1-v1.xlsx
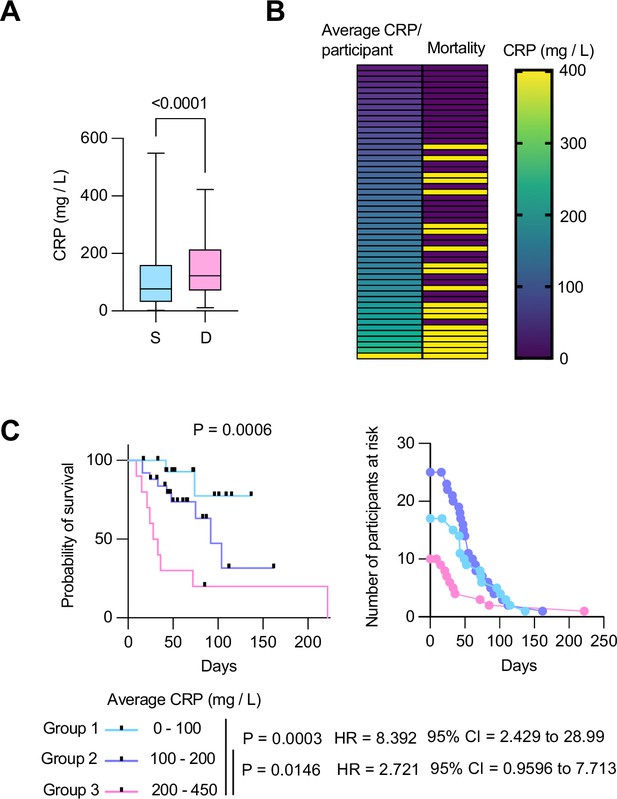
Longitudinal C-reactive protein (CRP) predicts survival probability in severe COVID-19 pneumonia.
(A) Individual CRP concentrations in 465 plasmas from 63 participants with maximum WHO severity grade 7 COVID-19 pneumonia segregated into survivors (n=43) and deceased (n=20) groups from the Berlin COVID-19 study. (B) Participants ordered by their longitudinal average CRP concentrations shown in the left column. Mortality is depicted in yellow in the right column. (C) Kaplan Meier survival probabilities (left panel) and numbers at risk (right panel) for patients segregated into three categories of longitudinal average CRP ranges: 0–100 mg / L (n=17), 100–200 mg / L (n=25), and 200–450 mg / L (n=10). Statistical significance (P), Hazard ratios (HR) and 95% confidence intervals (95% CI) for group 1 against group 3 and group 2 against group 3 are shown below the survival plot. Statistics by Mann-Whitney and Mantel-Cox log rank tests.
-
Figure 2—source data 1
C-reactive protein (CRP) concentrations, participant stratification, and survival analysis of patients in the Berlin COVID-19 study.
- https://cdn.elifesciences.org/articles/87030/elife-87030-fig2-data1-v1.xlsx
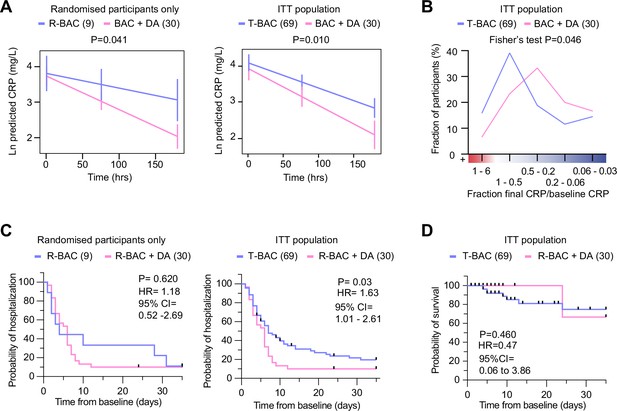
Analysis of primary and clinical endpoints.
(A) Fitted mean (95% confidence interval) from mixed model of natural log (C-reactive protein, CRP) over 7 days follow-up as the outcome. (Left panel) randomized participants only: Blue: participants randomized to R-BAC, n=9; Pink: participants randomized to R-BAC +DA, n=30. (Right panel) ITT population. Blue: T-BAC (CC-BAC and R-BAC) n=69; Pink: R-BAC +DA, n=30. Results were adjusted for natural log baseline CRP, age, sex, BMI, serious comorbidity (diabetes, cardiovascular disease, or hypertension), time and a treatment × time interaction. P-value generated by comparing least-square means between the arms. (B) Distribution of participants based on the change in CRP measured as a ratio of the final CRP reading within the 7 day treatment period over the baseline CRP reading per participant. Statistical analysis by Fisher’s test. (C) Kaplan-Meier plot showing time to discharge from hospital from baseline. Hazard ratio from Cox proportional hazards model adjusted for baseline CRP, age, sex, BMI, serious comorbidity. p-value from log-rank test. Blue: CC-BAC and participants randomized to R-BAC, n=69. Pink: participants randomized to R-BAC +DA, n=30. (D) Kaplan-Meier plot showing time to death over 35 days follow up. Hazard ratio from Cox proportional hazards model adjusted for baseline CRP, age, sex, BMI, serious comorbidity. p-value from log-rank test.
-
Figure 3—source data 1
Mean C-reactive protein (CRP), participant distribution by CRP change, probability of hospitalization and survival for the randomized and intention-to-treat (ITT) populations.
- https://cdn.elifesciences.org/articles/87030/elife-87030-fig3-data1-v1.xlsx
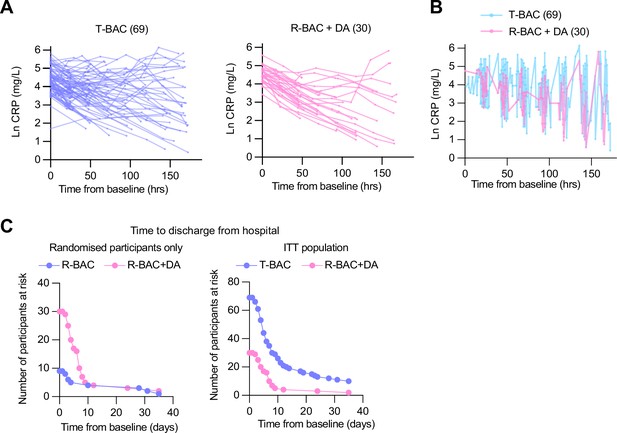
Primary and clinical endpoints.
(A) (Left panel) Natural log C-reactive protein (CRP) in best available care (BAC) (CC and randomized participants; blue). (Right panel) Natural log CRP in participants randomized to BAC+DA (pink). (B) Graph depicting the periodicity and frequency of blood sample collection for all post-baseline CRP values from contemporary control and randomized BAC (blue) or BAC+dornase alfa (BAC+DA, pink) patients pooled into a single timeline. (C). (Left panel) Numbers at risk for Kaplan-Meier plots of the time to discharge from hospital (Figure 3C). intention-to-treat (ITT) population (Blue: CC and participants randomized to BAC, n=69. Pink: participants randomized to BAC+DA, n=30). (Right panel) Numbers at risk depicting the time to discharge from hospital from baseline. Blue: participants randomized to R-BAC, n=9; Pink: participants randomized to R-BAC+DA, n=30. (Right panel) ITT population: Blue: T-BAC (CC-BAC and R-BAC) n=69; Pink: R-BAC+DA, n=30. Hazard ratio from Cox proportional hazards model adjusted for baseline CRP, age, sex, BMI, serious co-mor bidity (diabetes, cardiovascular disease, or hypertension).
-
Figure 3—figure supplement 1—source data 1
Individual C-reactive protein (CRP) readings and numbers at risk for length of hospitalization.
- https://cdn.elifesciences.org/articles/87030/elife-87030-fig3-figsupp1-data1-v1.xlsx
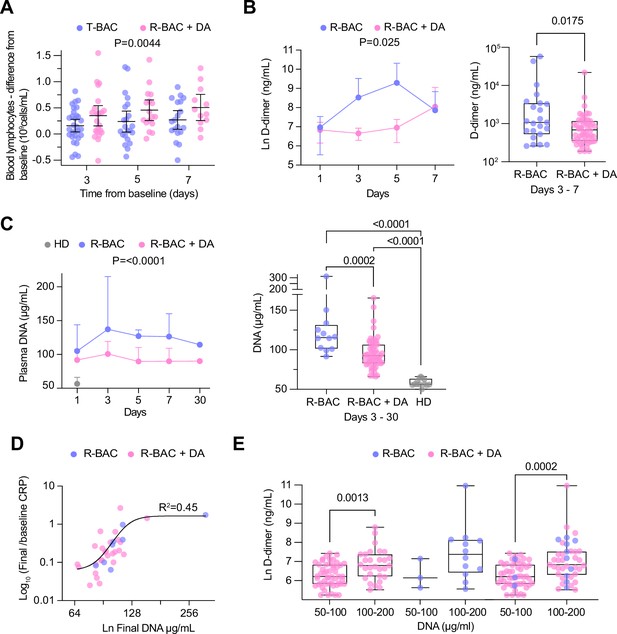
Analysis of secondary and exploratory endpoints in blood.
(A) Difference between the lymphocyte count for each day of the treatment period and the baseline in each intention-to-treat (ITT) participant who exhibited lymphopenia at baseline (<1 × 109 lymphocytes/mL). T-BAC (n=71 samples); R-BAC + DA (n=52 samples). The mean and 95% CI interval are shown with statistical analysis by two-way Anova. (B) (Left panel) Mean blood D-dimer levels per day in randomized R-BAC (blue) and R-BAC +DA (pink) participants with error bars depicting 95% CI. Statistical difference by mixed effects Anova analysis. (Right panel) D-dimer concentration in the randomized participant post-baseline blood samples from the R-BAC (n=11 samples) and R-BAC +DA (n=28 samples) groups. Statistical analysis by two-tailed unpaired parametric t-test. (C) (Left panel) Mean cell free (cf)-DNA levels per day in randomized R-BAC (n=22 samples, blue) and R-BAC +DA (n=89 samples, pink) participants, with error bars depicting standard deviation. Statistical analysis by mixed effects Anova. (Right panel) Pooled cf-DNA concentration measurements in post-baseline blood samples of R-BAC (n=12) and R-BAC +DA (n=59) groups from days 3, 7 and 30. Healthy donor plasma cf-DNA concentrations (HD, n=13 samples, grey) are shown for comparison. Statistical analysis by one-way Anova. (D) Correlation between the final cf-DNA levels and ratio of CRP at day-7 normalized to the baseline C-reactive protein (CRP) (CRPfinal/CRPbaseline) per randomized participant (Total: n=34; R-BAC: n=7, R-BAC +DA: n=27) . Fitting by non-linear regression. (E) Correlation between D-dimer and cf-DNA levels in the blood of participants randomized to R-BAC (blue) or to R-BAC +DA (DA) (pink), where samples were segregated depending on whether the corresponding levels of cf-DNA were <100μg/mL (R-BAC: n=3; R-BAC+ DA: n=51) or >100 μg/mL (R-BAC: n=12; R-BAC+ DA: n=29). Statistical analysis by unpaired parametric t-test.
-
Figure 4—source data 1
Change in blood lymphocytes, D-dimer, DNA and correlations between DNA and D-dimer in randomized participants.
- https://cdn.elifesciences.org/articles/87030/elife-87030-fig4-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | λ DNA (cIind 1 ts857 Sam 7) | Invitrogen | 25250010 | |
| Commercial assay or kit | Quant-iT PicoGreen dsDNA Reagent | Invitrogen | P7581 | |
| Chemical compound, drug | Histopaque-1119 | Sigma-Aldrich | 11191 | Density media for cell isolation |
| Chemical compound, drug | Percoll | Cytiva | 17544502 | Density media for cell isolation |
| Chemical compound, drug | HBSS without calcium, magnesium, phenol red | Cytiva | SH30588.01 | Balanced salt solution |
| Chemical compound, drug | 1 M HEPES | Sigma-Aldrich | SRE0065 | Biological buffer |
| Chemical compound, drug | Tris-EDTA | Sigma-Aldrich | 93283 | Biological buffer |
Patient baseline characteristics.
Age, gender, BMI, comorbidity frequency, CRP, WHO ordinal severity score, blood leukocyte counts, and blood procalcitonin and D-dimer concentrations at baseline for all participants (R-BAC, CC-BAC, and R-BAC +DA).
| R-BAC +DA (n=30) | R-BAC (n=9) | CC-BAC(n=60) | T-BAC (n=69) | Total (n=99) | |
|---|---|---|---|---|---|
| Age (years) | |||||
| Mean | 56.8 | 53.3 | 57.3 | 56.8 | 56.8 |
| SD | 12.5 | 13.7 | 14.5 | 14.3 | 13.7 |
| Median | 58.0 | 53.0 | 57.0 | 57.0 | 57.0 |
| Min | 32.0 | 31.0 | 23.0 | 23.0 | 23.0 |
| Max | 77.0 | 76.0 | 86.0 | 86.0 | 86.0 |
| Gender | |||||
| Female N (%) | 7 (23.3) | 2 (22.2) | 15 (25.0) | 17 (24.6) | 24 (24.2) |
| Male N (%) | 23 (76.7) | 7 (77.8) | 45 (75.0) | 52 (75.4) | 75 (75.8) |
| BMI (kg/m2) | |||||
| Mean | 27.8 | 30.8 | 27.8 | 28.2 | 28.0 |
| SD | 4.7 | 7.8 | 5.6 | 6.0 | 5.6 |
| Median | 26.5 | 28.9 | 27.9 | 28.2 | 27.7 |
| Min | 20.7 | 22.6 | 16.3 | 16.3 | 16.3 |
| Max | 41.7 | 48.4 | 43.8 | 48.4 | 48.4 |
| Baseline CRP (mg/L) | |||||
| Mean | 101.9 | 91.9 | 100.7 | 99.5 | 100.2 |
| SD | 52.2 | 68.1 | 68.3 | 67.8 | 63.3 |
| Median | 86.3 | 74.6 | 75.8 | 75.3 | 79.6 |
| Min | 25.2 | 18.9 | 30.8 | 18.9 | 18.9 |
| Max | 261.5 | 221.6 | 336.4 | 336.4 | 336.4 |
| Comorbidity | |||||
| No N (%) | 16 (53.3) | 3 (33.3) | 28 (46.7) | 31 (44.9) | 47 (47.5) |
| Yes N (%) | 14 (46.7) | 6 (66.7) | 32 (53.3) | 38 (55.1) | 52 (52.5) |
| WHO ordinal severity score | |||||
| Mean | 5.0 | 5.0 | 4.63 | - | - |
| SD | 0.0 | 0.5 | 1.33 | - | - |
| Median | 5.0 | 5.0 | 5 | - | - |
| Min | 5.0 | 4.0 | 3 | - | - |
| Max | 5.0 | 6.0 | 7 | - | - |
| WBC count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 6.7 | 7.0 | 10.6 | 10.2 | 9.1 |
| SD | 2.5 | 2.7 | 9.2 | 8.7 | 7.6 |
| Median | 6.5 | 7.0 | 9.5 | 8.9 | 7.9 |
| Min | 3.1 | 1.8 | 1.8 | 1.8 | 1.8 |
| Max | 12.9 | 10.3 | 72.6 | 72.6 | 72.6 |
| Neutrophil count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 5.7 | 5.6 | 9.1 | 8.7 | 7.8 |
| SD | 2.3 | 2.6 | 8.8 | 8.4 | 7.2 |
| Median | 5.3 | 5.8 | 7.9 | 7.9 | 6.7 |
| Min | 2.4 | 1.2 | 1.2 | 1.2 | 1.2 |
| Max | 10.9 | 8.6 | 69.5 | 69.5 | 69.5 |
| Lymphocyte count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 0.7 | 0.9 | 0.9 | 0.9 | 0.9 |
| SD | 0.3 | 0.4 | 0.5 | 0.5 | 0.5 |
| Median | 0.5 | 0.9 | 0.8 | 0.8 | 0.7 |
| Min | 0.2 | 0.4 | 0.1 | 0.1 | 0.1 |
| Max | 1.5 | 1.5 | 3.7 | 3.7 | 3.7 |
| Monocyte count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 0.4 | 0.4 | 0.5 | 0.5 | 0.4 |
| SD | 0.2 | 0.3 | 0.3 | 0.3 | 0.3 |
| Median | 0.3 | 0.3 | 0.4 | 0.4 | 0.4 |
| Min | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Max | 0.9 | 0.8 | 1.7 | 1.7 | 1.7 |
| Eosinophil count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| SD | 0.1 | 0.0 | 0.1 | 0.1 | 0.1 |
| Median | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Min | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Max | 0.2 | 0.1 | 0.6 | 0.6 | 0.6 |
| Basophil count (×109 /L) | |||||
| N | 30 | 9 | 60 | 69 | 99 |
| Mean | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| SD | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Median | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Min | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Max | 0.2 | 0.1 | 0.1 | 0.1 | 0.2 |
| Procalcitonin count (ng/ml) | |||||
| N | 27 | 8 | 1 | 9 | 36 |
| Mean | 0.3 | 0.3 | 19.3 | 2.4 | 0.8 |
| SD | 0.4 | 0.3 | - | 6.3 | 3.2 |
| Median | 0.1 | 0.2 | 19.3 | 0.2 | 0.2 |
| Min | 0.1 | 0.1 | 19.3 | 0.1 | 0.1 |
| Max | 1.8 | 0.8 | 19.3 | 19.3 | 19.3 |
| D-dimer (ug/L) FEU | |||||
| N | 30 | 9 | 15 | 24 | 54 |
| Mean | 885.0 | 909.1 | 1059.3 | 1003.0 | 937.4 |
| SD | 1154.5 | 1054.1 | 1115.0 | 1071.8 | 1109.6 |
| Median | 545.0 | 570.0 | 600.0 | 585.0 | 570.0 |
| Min | 190.0 | 1.9 | 280.0 | 1.9 | 1.9 |
| Max | 6580.0 | 3570.0 | 4460.0 | 4460.0 | 6580.0 |
Primary endpoint and sensitivity analysis.
Mean CRP concentrations over 7 days follow up from baseline for CC-BAC, R-BAC, and R-BAC +DA participants and different comparisons between the three groups as a whole, or stratified by whether in addition to dexamethasone they also received treatment with either remdesivir or tocilizumab at baseline and during the course of the assessment. The difference between the mean log CRP with 95% confidence interval (95% CI) and statistical test values in the group comparisons are shown.
| CRP (mg/L) | BAC+DA | BAC | Difference | p-value* |
|---|---|---|---|---|
| ITT population (R-BAC+DA, R-BAC, CC-BAC) | ||||
| N | 30 | 69 | ||
| LS means log(CRP)* (95% CI) | 3.15 (2.87–3.42) | 3.55 (3.35–3.75) | –0.4 (–0.71 to –0.10) | 0.010 |
| LS means CRP† (95% CI) | 23.23 (17.71–30.46) | 34.82 (28.55–42.47) | 0.67 (0.49–0.91) | |
| Sensitivity Analyses | ||||
| PP population (R-BAC+DA, R-BAC, CC-BAC) | ||||
| N | 29 | 68 | ||
| LS means log(CRP)* (95% CI) | 3.12 (2.85–3.39) | 3.55 (3.36–3.74) | –0.43 (–0.73 to –0.13) | 0.006 |
| LS means CRP† (95% CI) | 22.64 (17.35–29.54) | 34.82 (27.7–42.21) | 0.65 (0.48–0.88) | |
| ITT population (R-BAC+DA, R-BAC) | ||||
| N | 30 | 9 | ||
| LS means log(CRP)* (95% CI) | 3.1 (2.84–3.35) | 3.59 (3.13–4.06) | –0.5 (–0.97 to –0.02) | 0.041 |
| LS means CRP† (95% CI) | 22.12 (17.16–28.5) | 36.34 (22.79–57.94) | 0.61 (0.38–0.98) | |
| ITT population (R-BAC +DA, CC-BAC) | ||||
| N | 30 | 60 | ||
| LS means log(CRP)* (95% CI) | 3.18 (2.91–3.45) | 3.56 (3.35–3.76) | –0.37 (–0.68 to –0.06) | 0.019 |
| LS means CRP† (95% CI) | 24.09 (18.36–31.6) | 35.03 (28.44–43.15) | 0.69 (0.5–0.94) | |
| ITT population (R-BAC, CC-BAC) | ||||
| N | 9 | 60 | ||
| LS means log(CRP)* (95% CI) | 3.78 (3.23–4.33) | 3.53 (3.3–3.77) | 0.24 (−0.32–0.8) | 0.386 |
| LS means CRP† (95% CI) | 43.69 (25.18–75.8) | 34.23 (27.11–43.24) | 1.28 (0.73–2.23) | |
| Area under the log(CRP), standardized by days followed up, over 7 days follow-up ITT population (R-BAC +DA, R-BAC, CC-BAC) | ||||
| N | 30 | 69 | ||
| LS means area ‡ (95% CI) | 3.45 (3.22–3.68) | 3.72 (3.55–3.88) | –0.27 (–0.53 to –0.01) | 0.043 |
| ITT population (R-BAC +DA, R-BAC, CC-BAC) including last pre-dexamethasone CRP value | ||||
| N | 30 | 69 | ||
| LS means log(CRP)* (95% CI) | 3.16 (2.83–3.5) | 3.69 (3.44–3.93) | –0.53 (–0.91 to –0.14) | 0.007 |
| LS means CRP† (95% CI) | 23.57 (16.85–32.970) | 39.92 (31.32–50.89) | 0.59 (0.4–087) | |
| ITT population (R-BAC +DA, R-BAC, CC-BAC) stratified by BAC treatment | ||||
| No remdesivir or tocilizumab | ||||
| N | 12 | 39 | ||
| LS means log(CRP)* (95% CI) | 3.29 (2.83–3.76) | 3.75 (3.45–4.04) | –0.45 (−0.96–0.05) | 0.079 |
| LS means CRP† (95% CI) | 26.97 (16.87–43.11) | 42.35 (31.44–57.04) | 0.64 (0.38–1.06) | |
| Remdesivir no tocilizumab | ||||
| N | 16 | 23 | ||
| LS means log(CRP)* (95% CI) | 3.16 (2.79–3.53) | 3.5 (3.18–3.83) | –0.35 (−0.79–0.1) | 0.123 |
| LS means CRP† (95% CI) | 23.53 (16.29–33.99) | 33.26 (23.97–46.15) | 0.71 (0.45–1.1) | |
| Tocilizumab no remdesivir | ||||
| N | 1 | 5 | ||
| Remdesivir and tocilizumab | ||||
| N | 1 | 2 | ||
-
*
From linear repeated measures model, adjusted for natural log(baseline CRP, age, sex, BMI, serious condition, time, treatment, a treatment*time interaction, and subject as a random effect. Least squares means compared at mean follow-up time).
-
†
Antilog of estimates from *Ratio of BAC +dorna-alfa: BAC shown in the difference column.
-
‡
From linear model, adjusted for natural log(baseline CRP, age, sex, BMI, serious condition, and treatment).
Primary endpoint by day.
Mean Log CRP concentrations and standard deviation (SD) for each day over 7 days follow-up by treatment for the intention-to-treat (ITT) population including all individuals (R-BAC +DA, R-BAC, CC-BAC).
| Days from baseline | MeanT-BAC +DA | SDT-BAC +DA | NT- BAC +DA | MeanR-BAC | SDR-BAC | NR-BAC |
|---|---|---|---|---|---|---|
| 1 | 4.058 | 0.591 | 21 | 4.029 | 0.734 | 42 |
| 2 | 3.538 | 0.658 | 33 | 3.852 | 0.937 | 52 |
| 3 | 3.332 | 0.816 | 24 | 3.839 | 1.037 | 53 |
| 4 | 2.917 | 1.017 | 17 | 3.573 | 1.132 | 37 |
| 5 | 2.52 | 1.118 | 15 | 3.872 | 1.157 | 24 |
| 6 | 3.059 | 1.508 | 12 | 3.547 | 1.516 | 26 |
| 7 | 3.386 | 1.824 | 9 | 3.504 | 1.451 | 27 |
| 8 | 3.428 | - | 1 | 2.478 | 1.393 | 9 |
Additional files
-
Supplementary file 1
Supplementary tables.
(A) Dexamethasone administration prior to recruitment Duration of dexamethasone treatment prior to recruitment and initiation of dornase alfa treatment in randomized and contemporary control participants. (B) Secondary endpoints in randomized participants only: Time to discharge, D-dimer, lymphocyte counts and procalcitonin measurements. (C) Secondary clinical endpoints in in randomized and contemporary control participants: Admission to ICU rates, length of stay in ICU, time on oxygen over 7- and 35 days follow-up, duration of mechanical ventilation, and proportion of individuals with superadded bacterial pneumonia. (D) Safety Table depicting the reported adverse events, the degree of severity and the relationship to dornase alfa therapy. (E) Cumulative Summary Tabulations of Serious Adverse Events. Serious adverse effects in randomized R-BAC and R-BAC +DA participants separated by system order class (infections, respiratory, thoracic and mediastinal disorders and vascular disorders). A total of 6 events were reported, with 2 in the R-BAC and 4 in the R-BAC +DA groups.
- https://cdn.elifesciences.org/articles/87030/elife-87030-supp1-v1.docx
-
Supplementary file 2
Consort checklist and protocol.
- https://cdn.elifesciences.org/articles/87030/elife-87030-supp2-v1.docx
-
Supplementary file 3
Statistical analysis plan.
- https://cdn.elifesciences.org/articles/87030/elife-87030-supp3-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87030/elife-87030-mdarchecklist1-v1.docx





