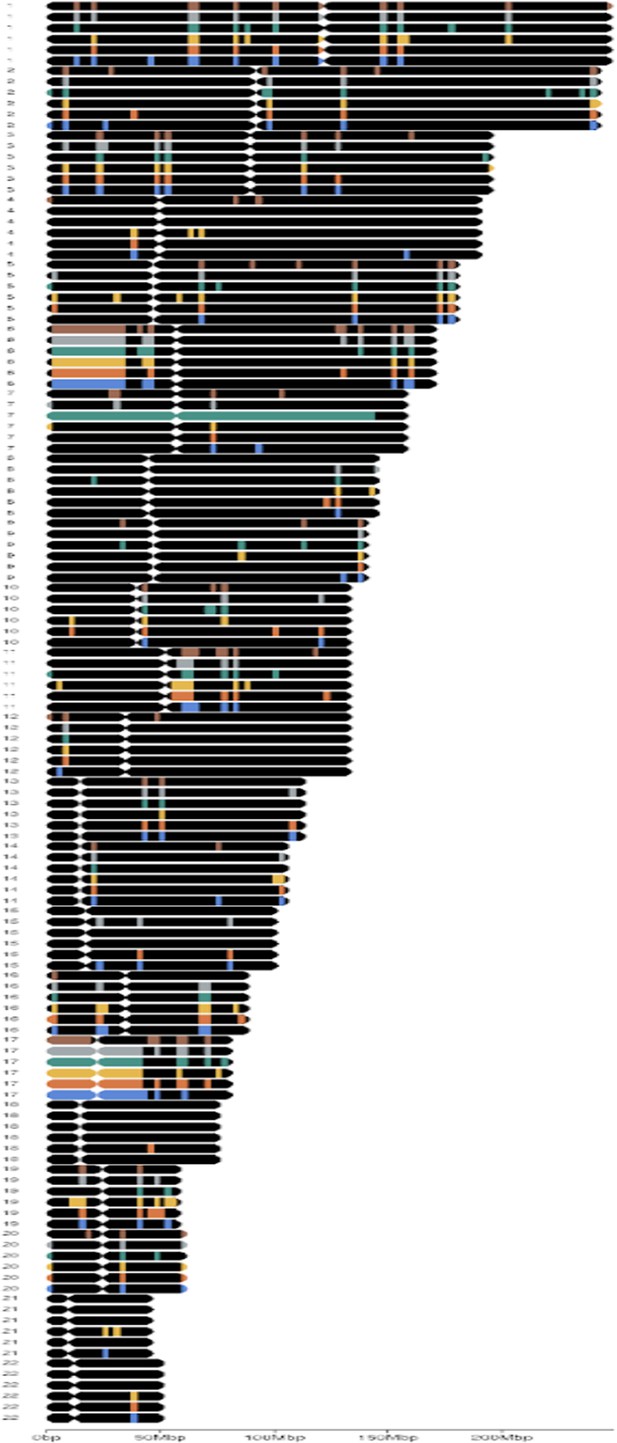
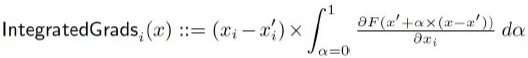Spatial transformation of multi-omics data unlocks novel insights into cancer biology
Figures
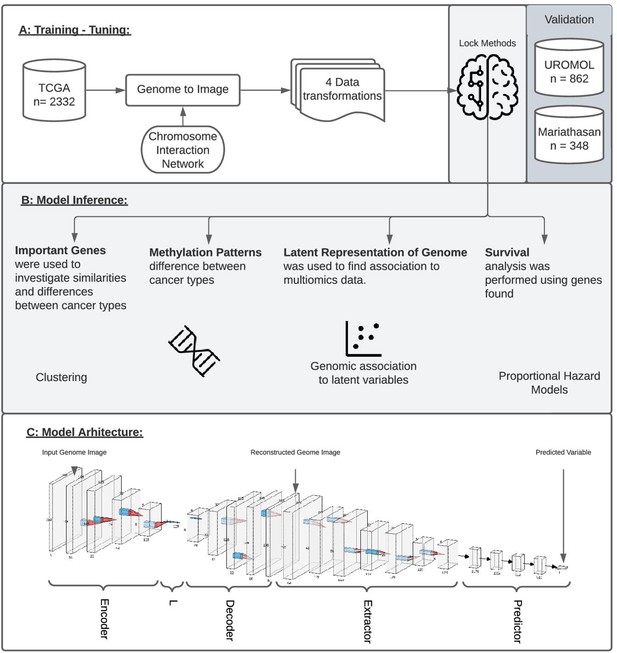
Study overview.
(A) The study utilized 2332 tumor samples representing six cancer types (bladder, uterine, stomach, ovarian, kidney, and colon) and transformed multi-omics data into images based on chromosome interaction networks. After the model was trained, we validated found genes with two independent cohorts representing early-stage bladder carcinoma (BLCA; UROMOL) and late-stage BLCA (Mariathasan). (B) The validation included looking at the most important genes driving metastatic disease, similar/different methylation patterns between cancer types, latent representation of genome data and looking at survival data. (C) The model architecture where the first part of the network encodes genome data into latent vector, L, followed by decoding where image is reconstructed. Next layers aim to extract information from the reconstructed image, concat it with L and make a final prediction.
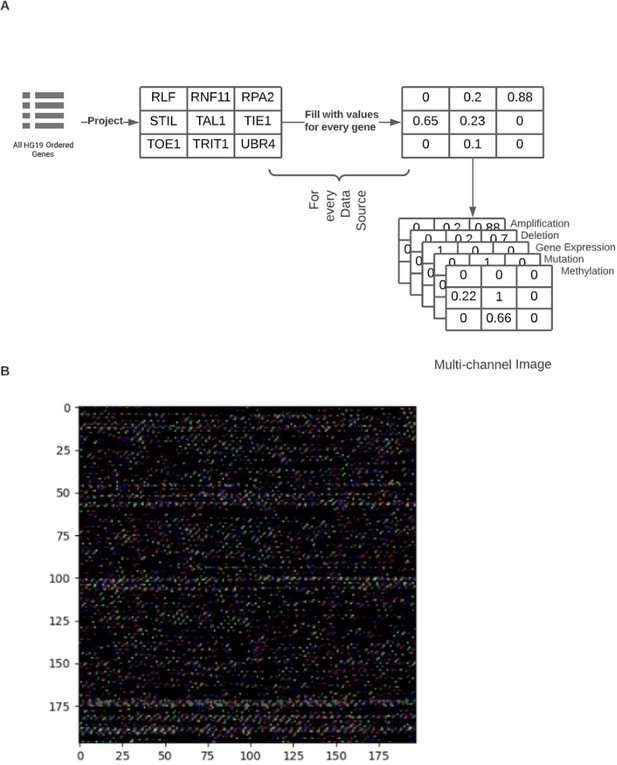
Figure representing an example of genome image construction.
(A) HG19 genes were ordered by chromosome and chromosome position. Next, based on gene position, the matrix was filled, where each cell represents a single gene. This was used for a template when creating matrices for every data source included in genome image construction. (B) Example of genome image.
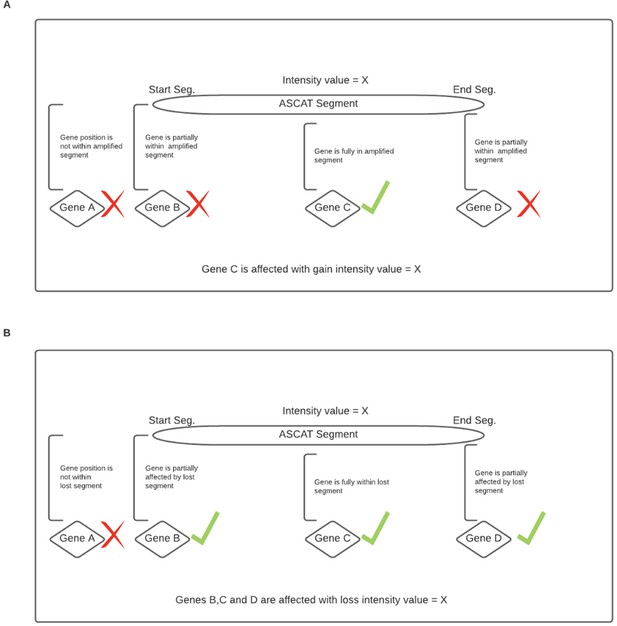
Overall representation of a process where chromosomal instability information was included in genome image construction.
(A) Amplification of a gene was declared when the gene was fully within an amplified segment. (B) Deletion of a gene was declared when the gene was partially within a deleted segment.
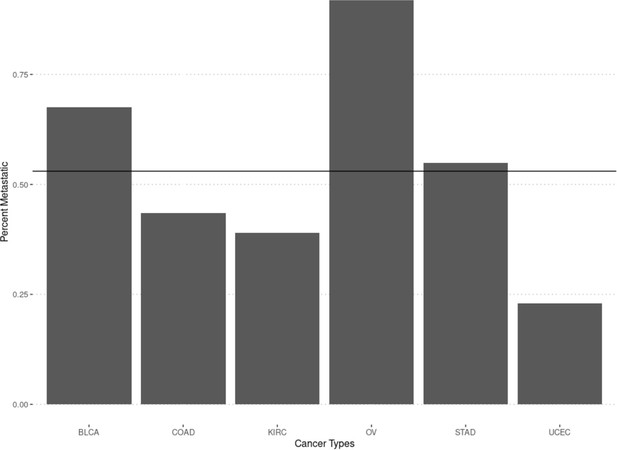
Figure showing the percent of samples classified as metastatic when using the stage as a variable.
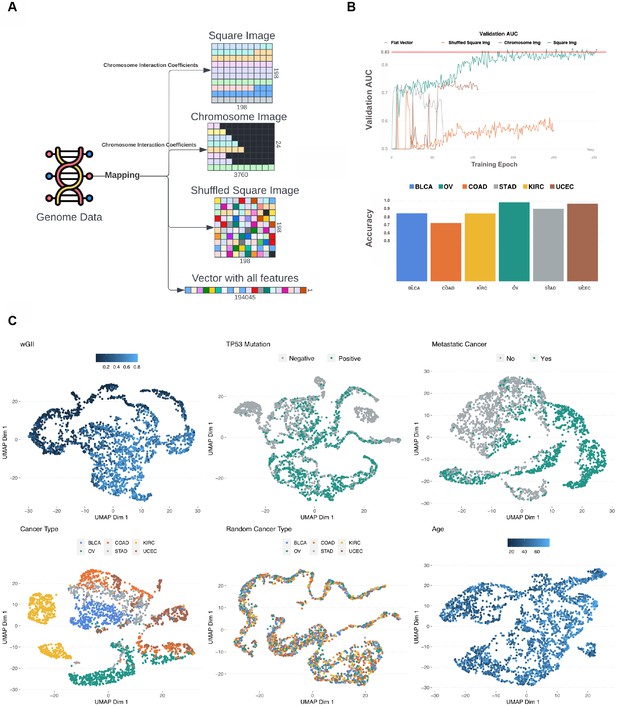
Data transformation overview.
(A) The multi-omics genome data was transformed into four image types: square image organized by chromosome interaction network, chromosome image organized by chromosome interaction network, randomly organized image, and flat vector containing all multi-omics data. (B) The x-axis represents epochs and the y-axis represents area under the curve (AUC) score of fixed 25% data we used for accuracy assessment within the TCGA cohort. All four image types were used in training for metastatic disease prediction and the square image organized by chromosome interaction network resulted in best model performance (green color). The red line shows where the model resulted in the best loss. All curves stopped when the loss started increasing, indicating overfitting. The bar plot shows the proportion of correctly predicted (metastatic disease) in every cancer type included in the study. (C) Two-dimensional representation of vector L using Uniform Manifold Approximation and Projection (UMAP) for each predicted variable. Colors indicate the output variable which was used in the specific run.
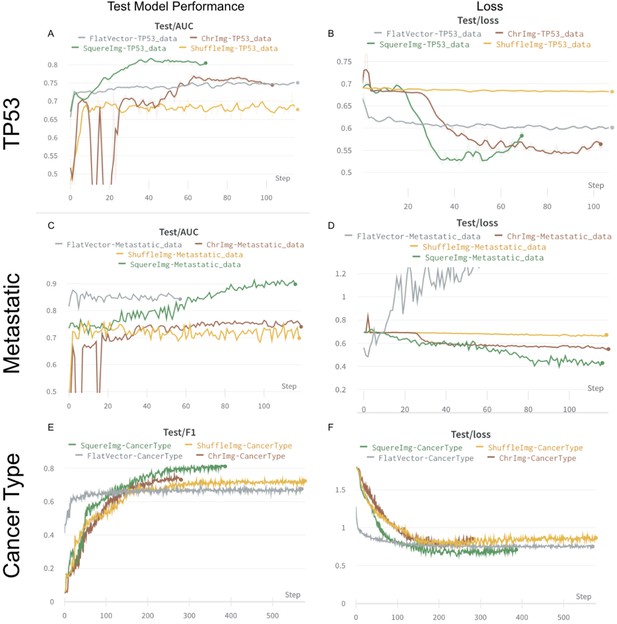
Figure showing the process of training four image transformations in classification scenarios.
x-axis represents epochs and y-axis represents area under the curve (AUC)/F1 (panels A, C, E). On panels B, D, F, y-axis is switched with loss.
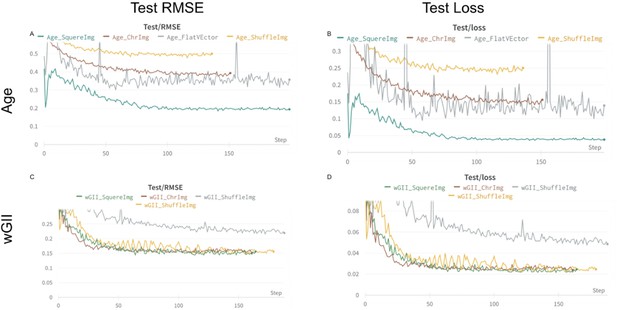
Figure showing the process of training four image transformations in regression scenarios.
x-axis represents epochs and y-axis represents Root Mean Squared Error (RMSE). (A) Panel shows RMSE in test cohort when training for Age. (B) Panel shows loss function in test cohort when training for Age. (C) Panel shows RMSE in test cohort when training for weighted genome integrity intex (wGII) . (D) Panel shows loss function in test coohort when training for wGII.
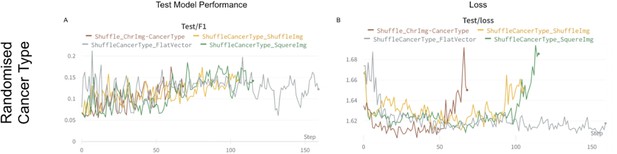
Figure showing the process of training four image transformations in classification scenarios (negative control).
x-axis represents epochs and y-axis represents F1 (panel A). On panel B, y-axis is switched with loss.
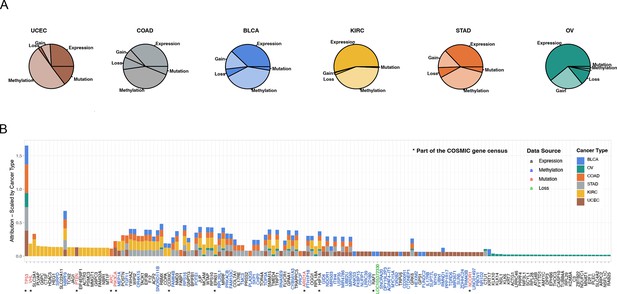
The most important events in metastatic disease development.
(A) Pieplot showing the relative importance of each data source when predicting metastatic disease for each cancer type included in the study. (B) Top 50 genes for every cancer type scale by cancer type. The star symbol below the gene names indicates that the gene is part of COSMIC gene consensus. The color of the gene name indicates the data source and color of the bar indicates the cancer type.
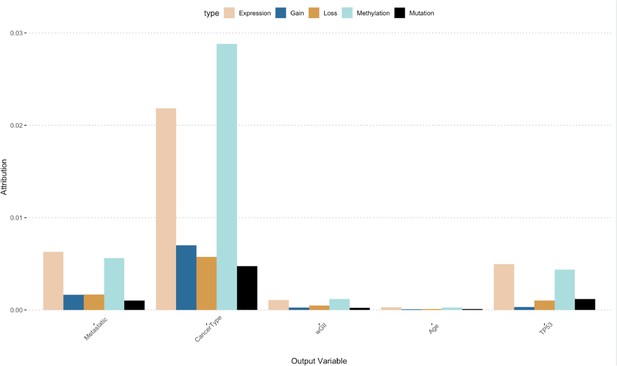
The figure shows the relative attribution of each variable we predicted using the GENIUS framework.
The color represents the relative importance of a specific data source in a specific scenario (x-axis).
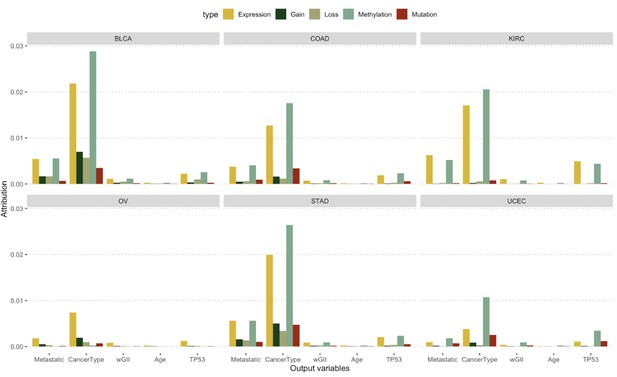
The figure shows the relative attribution of each variable we predicted using the GENIUS framework (x-axis) but split by cancer type.
The color represents the relative importance of a specific data source in a specific scenario (x-axis) and each panel represents cancer type.
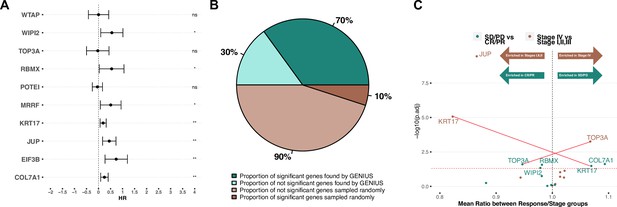
Validation on late-stage immunotherapy-treated bladder cancer (Mariathasan).
(A) Forest plot showing top 10 expressed/methylated genes in multivariate cox proportional hazard model. X-axis indicates Hazard Rate (HR). Stars indicates significance (* P < 0.05, ** P < 0.01, *** P < 0.001), "ns" indicates not significant. (B) Comparison of median percent of randomly selected genes versus genes picked by GENIUS in cox proportional hazard model. (C) Volcano plot showing top 10 expressed/methylated genes and their enrichment in two comparisons; stages I, II and III versus stage IV and immunotherapy response (CR: complete response, PR: partial response) versus no response (SD: stable disease, PD: progressive disease). Two genes show association in opposite directions, indicated by red lines (KRT17, associated with low stage and poor immunotherapy response, and TOP3A, associated with high stage and improved immunotherapy response).
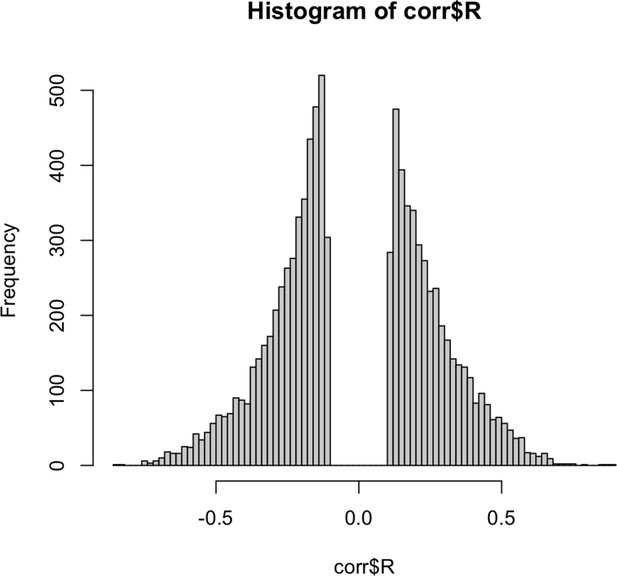
Histogram of correlation coefficients between each gene found in BLCA TCGA gene expression data and methylation data.
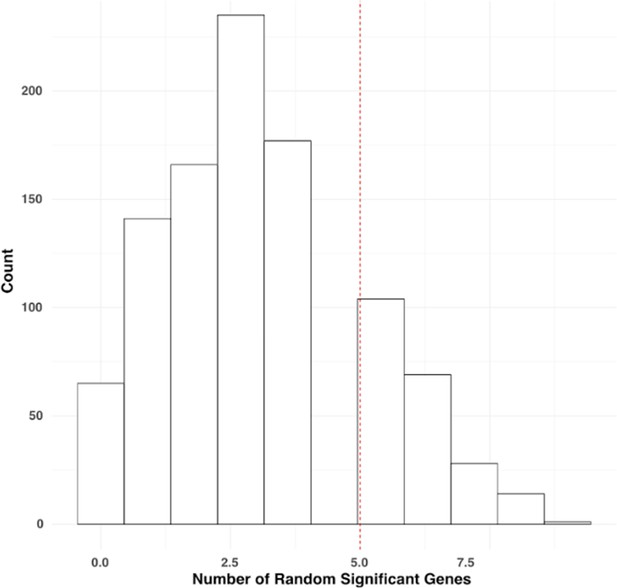
Histogram showing distribution of number of randomly picked significant genes in Mariathasan dataset.
Red line indicates the number of significant genes found based on the GENIUS analysis.
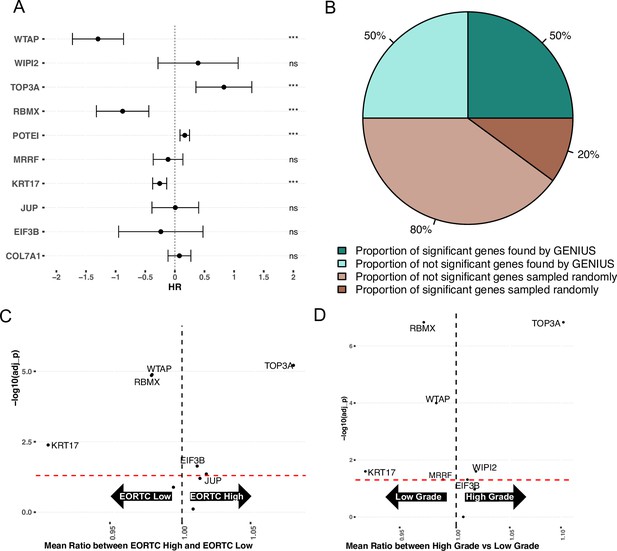
Validation on early-stage bladder cancer (UROMOL).
(A) Forest plot showing top 10 expressed/methylated genes picked by GENIUS for BLCA. X-axis indicates Hazard Rate (HR). Stars indicates significance (* P < 0.05, ** P < 0.01, *** P < 0.001), "ns" indicates not significant (B) Comparison of median percent of randomly selected genes versus genes picked by GENIUS in cox proportional hazard model. (C) Volcano plot showing association of the top 10 expressed/methylated genes relative to EORTC-Low and EORTC-High groups. (D) Volcano plot showing association of the top 10 expressed/methylated genes relative to low- and high-grade BLCA tumors.
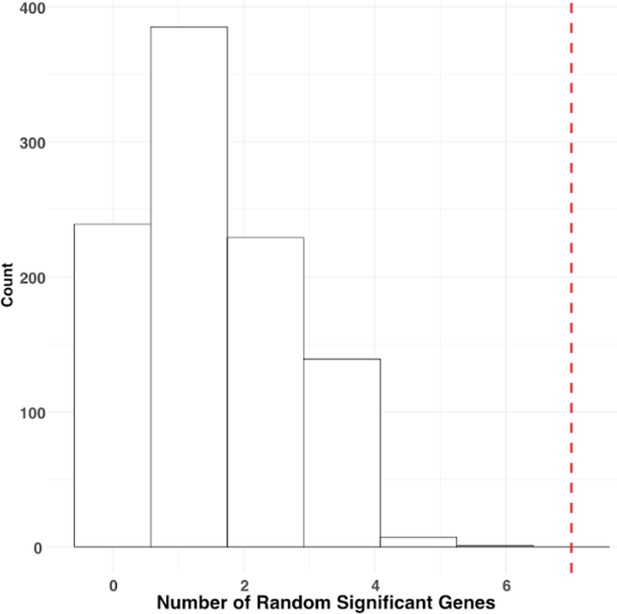
Histogram showing distribution of number of randomly picked significant genes in UROMOL dataset.
Red line indicates the threshold for significant results.
Tables
Summary of BLCA genes in two validation cohorts.
| Gene | Early stage | Late stage (immunotherapy) | Description |
|---|---|---|---|
| TOP3A | HR > 0 (PFS), high grade, high EORTC | HR = 0 (OS), enriched in stage IV, enriched in CR/PR | Catalyses the transient breaking and rejoining of a single strand of DNA, involved in regulation of recombination and homology-directed repair. Positive association to OS in OV (de Nonneville et al., 2022) |
| RBMX | HR < 0 (PFS), low grade, low EORTC | HR > 0 (OS), enriched in CR/PR | Associated with translational control and DNA damage pathways. Reported to be negatively correlated with tumor stage, histological grade, and poor patient prognosis in BLCA (Song et al., 2020) |
| POTEI | HR = 0 (PFS) | HR = 0 (OS) | POTE family of proteins is associated with apoptotic cells (Yu et al., 2023) |
| KRT17 | HR < 0 (PFS), low grade, low EORTC | HR > 0 (OS), enriched in stages I–III, enriched in SD/PD | Associated with structural molecule activity and MHC class II receptor activity. Associated with metastasis and angiogenesis in variety of tumor types (Ji et al., 2021) |
| WIPI2 | HR = 0 (PFS), high grade | HR > 0 (OS), enriched in CR/PR | Component of the autophagy machinery that controls the major intracellular degradation process. WIPI2 is suggested as a biomarker for predicting colorectal cancer prognosis (Yu et al., 2023) |
| MRRF | HR = 0 (PFS), low grade | HR > 0 (OS) | Associated with the ribosome recycling factor, which is a component of the mitochondrial translational machinery. High expression is associated with poor outcome in ovarian cancer (Song et al., 2020) |
| EIF3B | HR = 0 (PFS), high EORTC | HR > 0 (OS) | Eukaryotic translation initiation factor 3 subunit B is a promoter associated with pancreatic cancer (de Nonneville et al., 2022) |
| JUP | HR = 0 (PFS) | HR > 0 (OS), enriched in stages I–III | Common junctional plaque protein. Controversial role in different malignancies. Knockdown of JUP in epithelium-like GC cells causes EMT and promotes GC cell migration and invasion (Chen et al., 2021) |
| WTAP | HR <0 (PFS), low EORTC, low grade | HR = 0 (OS) | Wilms’ tumor 1-associating protein is associated to RNA methylation modifications, which regulate biological processes such as RNA splicing, cell proliferation, cell cycle, and embryonic development (Chen et al., 2021) |
| COL7A1 | HR = 0 (PFS) | HR > 0 (OS), enriched in SD/PD | Associated with metabolism of proteins and integrins in angiogenesis. Aberrant gene expression is associated with distinct tumor environment, metastasis and survival in multiple cancer types (Oh et al., 2021) |
Additional files
-
Supplementary file 1
Table shows the top 50 genes associated to metastatic disease development for every cancer type.
- https://cdn.elifesciences.org/articles/87133/elife-87133-supp1-v1.xlsx
-
Supplementary file 2
Summarized results for all output variables, containing top 10 events with respect to the predicted variable.
- https://cdn.elifesciences.org/articles/87133/elife-87133-supp2-v1.xlsx
-
Supplementary file 3
Top 10 expressed or methylated genes for every cancer type associated to metastatic disease.
- https://cdn.elifesciences.org/articles/87133/elife-87133-supp3-v1.xlsx
-
Supplementary file 4
Full model specifications for every multivariate cox proportional hazard model in late-stage immunotherapy-treated bladder cancer validation cohort (Mariathasan).
- https://cdn.elifesciences.org/articles/87133/elife-87133-supp4-v1.xlsx
-
Supplementary file 5
Full model specifications for every multivariate cox proportional hazard model in early-stage bladder cancer validation cohort (UROMOL).
- https://cdn.elifesciences.org/articles/87133/elife-87133-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87133/elife-87133-mdarchecklist1-v1.pdf