A tRNA modification in Mycobacterium tuberculosis facilitates optimal intracellular growth
Figures
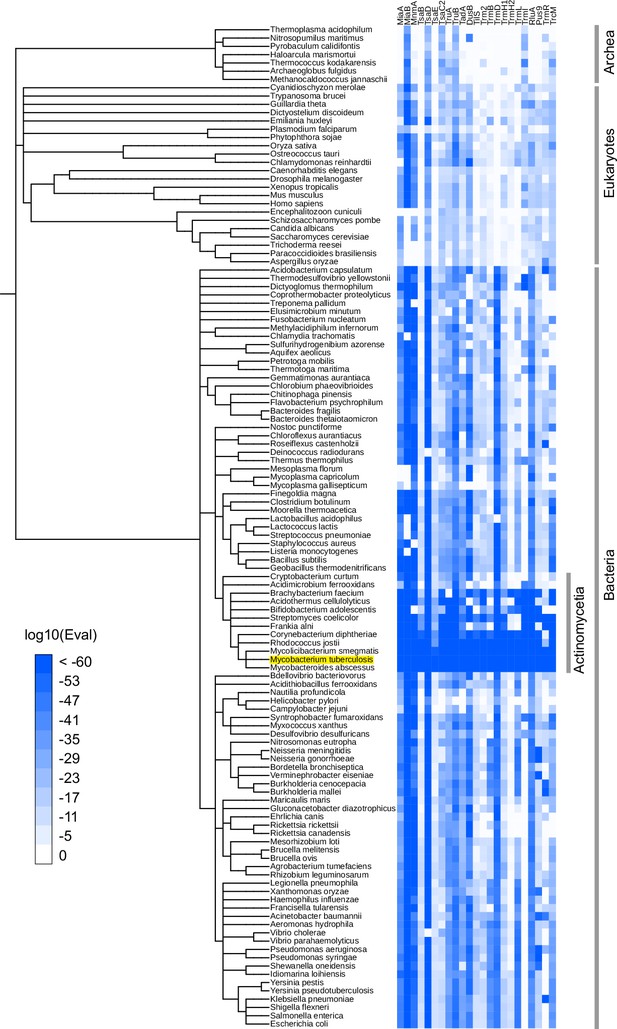
Phylogenetic distribution of Mtb tRNA modifying enzyme homologs.
Heatmap of log10 E-values from BLAST search results. BLAST searches were conducted against 120 manually picked organisms using Mtb tRNA modifying enzymes as queries. When one organism has multiple hits, the lowest log10(Eval) values among hits are shown. iTol (Letunic and Bork, 2021) was used to depict the results.
-
Figure 1—source data 1
log10 E-values from BLAST searching for the homologs of Mtb tRNA modifying enzymes in 120 organisms.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig1-data1-v1.xlsx

Heatmap of misincorporation and early termination frequency in sequencing of tRNAs from wild-ype Mtb.
Heatmaps show misincorporation (A) and termination (B) frequencies at all positions across tRNAs (read 5′ to 3′). Predicted modifications are labeled based on similarity to known modifications in other organisms and the presence of the tRNA modifying enzyme homologs (Supplementary file 3). The positions with more than 10% misincorporation in Mtb but not in E. coli are depicted in white in A. Representative data of two independent experiments with similar results are shown. (C) M. tuberculosis tRNA modifications predicted in this study. Schematic tRNA secondary structure with sites of modifications identified either by the presence of modifying enzymes and/or tRNA sequencing (tRNA-seq). Modifications and tRNA species that are not observed in E. coli are shown in red. Modifications and positions that are predicted by both RT-derived signature and the presence of the homologs of tRNA modifying enzymes are shown in yellow (without chemical treatment) and green (with chemical treatment), whereas modifications that are only predicted by the presence of the homologs are shown in light blue. Genes reported to be essential in Mtb are shown in bold.
-
Figure 2—source data 1
Misincorporation and early termination frequencies in sequencing of tRNAs from wild-type Mtb.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig2-data1-v1.xlsx
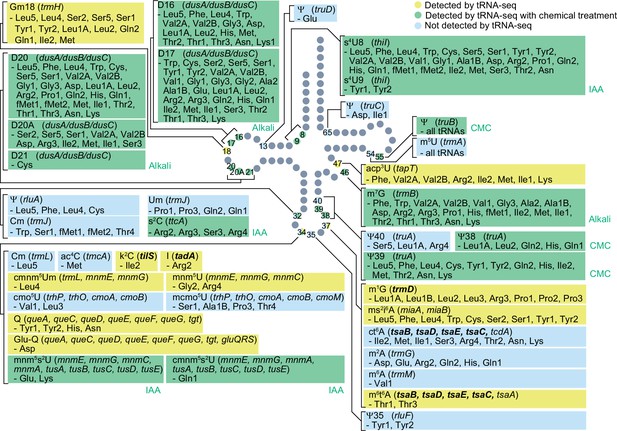
E. coli tRNA modifications detected by tRNA sequencing (tRNA-seq).
Schematic tRNA secondary structure with sites of modifications. Modifications and positions that are detected by RT-derived signature in yellow (without chemical treatment in Zhang et al., 2022) and green (with chemical treatment in this study), whereas modifications that are not detected by RT-derived signature are shown in light blue. Sites shown in yellow or green were not detected in all the listed tRNA species known to be modified. Genes reported to be essential in E. coli are shown in bold.
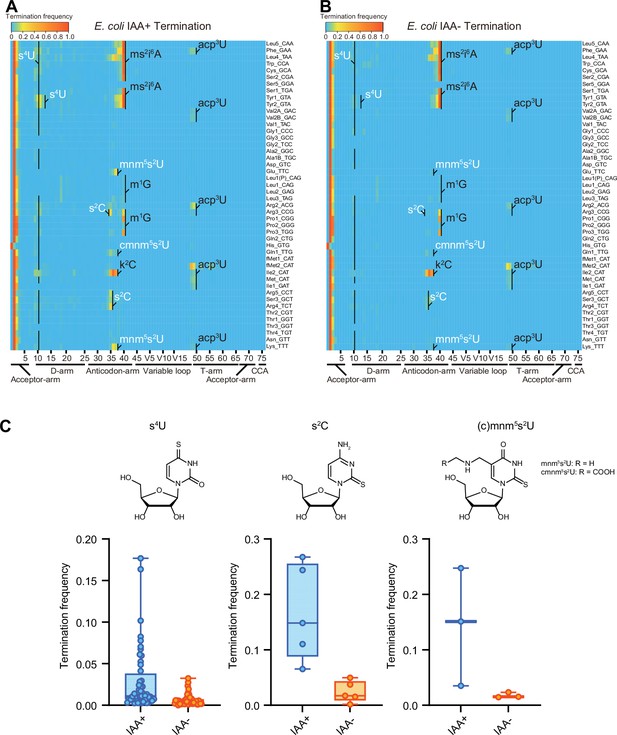
Iodoacetamide (IAA) treatment promotes detection of sulfur modifications on tRNAs by enhancing termination signals.
Heatmaps of the termination signals of E. coli tRNAs treated with (A) or without (B) IAA. Known modification sites, including sulfur modifications (s4U, s2C, s2U in white) are shown. (C) Termination frequency at s4U (n = 48), s2C (n = 5), and s2U (n = 3) sites of tRNAs treated with or without IAA. The experiment was performed once.
-
Figure 3—source data 1
Termination frequencies of E. coli tRNAs treated with or without iodoacetamide (IAA).
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig3-data1-v1.xlsx
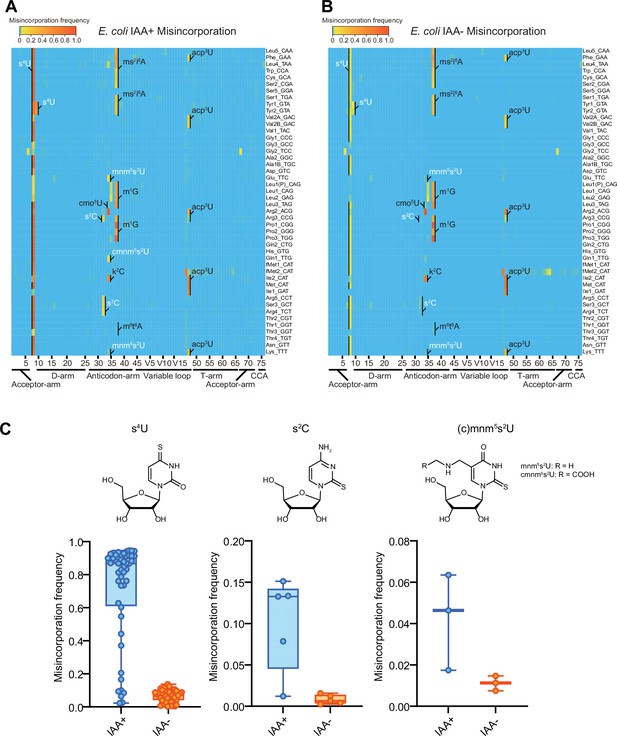
Iodoacetamide (IAA) treatment promotes detecting sulfur modifications by enhancing misincorporation signals.
Heatmaps of the misincorporation signals of E. coli tRNAs treated with (A) or without (B) IAA. Known modification sites, including sulfur modifications (s4U, s2C, s2U in white) are shown. (C) Misincorporation frequency at s4U (n = 48), s2C (n = 5), and s2U (n = 3) sites of tRNAs treated with or without IAA.
-
Figure 3—figure supplement 1—source data 1
Misincorporation frequencies of E. coli tRNAs treated with or without iodoacetamide (IAA).
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig3-figsupp1-data1-v1.xlsx

Heatmap and plot of early termination frequency from sequencing of tRNAs from wild-type and MtbΔmnmA with and without RNA alkylation.
Heatmap of early termination frequencies across tRNA molecules and positions for wild-type (WT) (A) and MtbΔmnmA (B). Sulfur modification is shown in white. (C) Plot of termination frequencies at position 37 in WT Mtb and MtbΔmnmA for lysine_UUU, glutamate_UUG, and glutamine_UUC isoacceptors (n = 3). IAA: iodoacetamide. The experiment was performed once.
-
Figure 4—source data 1
Termination frequencies from sequencing of tRNAs from wild-type and MtbΔmnmA with and without RNA alkylation.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig4-data1-v1.xlsx

1-Cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) and alkali treatment facilitate detection of ψ, D, and m7G modifications in E. coli.
Heatmaps of the misincorporation signals of E. coli tRNAs treated with (A) or without (B) CMC. In both conditions, tRNAs are incubated in alkali condition. Known Ψ, D, and m7G sites are shown. Ψ is shown in white. (C) Misincorporation frequency at known Ψ (n = 80), D (n = 76), and m7G (n = 25) sites of tRNAs treated with CMC+/alkali+ or CMC−/alkali+, and tRNAs without treatment. The experiment was performed once.
-
Figure 5—source data 1
Misincorporation frequencies from sequencing of E. coli tRNAs treated with or without 1-cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC).
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig5-data1-v1.xlsx
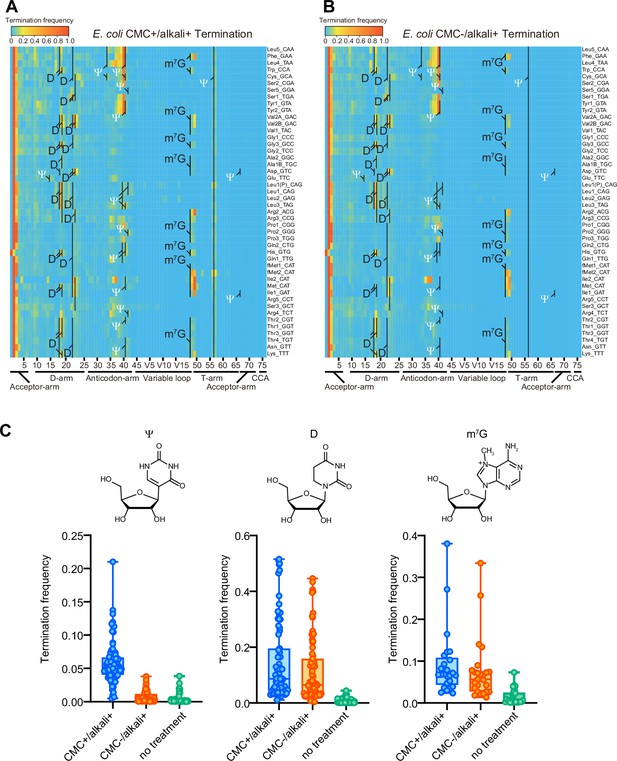
1-Cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) and the following alkali treatment facilitate detecting additional modifications.
Heatmaps of the termination signals of E. coli tRNAs treated with (A) or without (B) CMC. In both conditions, tRNAs are incubated at an alkali condition. Known Ψ (in white), D, and m7G sites are shown. (C) Termination frequency at known Ψ (n = 80), D (n = 75), and m7G (n = 25) sites of tRNAs treated with CMC+/alkali+ or CMC−/alkali+, and tRNAs without treatment.
-
Figure 5—figure supplement 1—source data 1
Termination frequencies from sequencing of E. coli tRNAs treated with or without 1-cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC).
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig5-figsupp1-data1-v1.xlsx
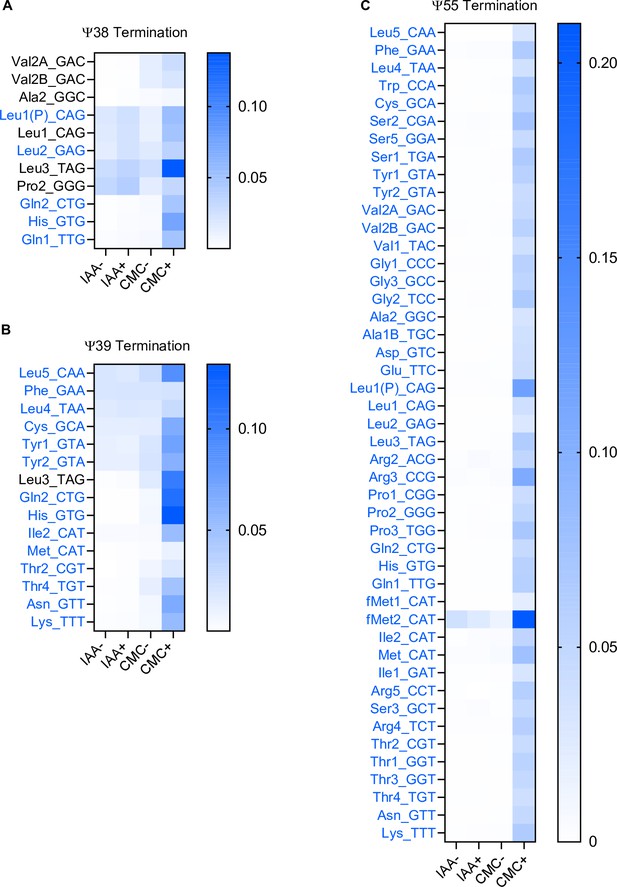
1-Cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) treatment facilitate detecting ψ.
Heatmaps of the termination frequencies at the uridine in E. coli tRNAs at positions 40 (A), 41 (B), and 57 (ψ55) (C). tRNA species where U at the indicated position is known to be ψ are shown in blue. Chemical treatment is shown in bottom.
-
Figure 5—figure supplement 2—source data 1
Termination frequencies at the uridine in E. coli tRNAs at positions 40, 41, and 57.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig5-figsupp2-data1-v1.xlsx
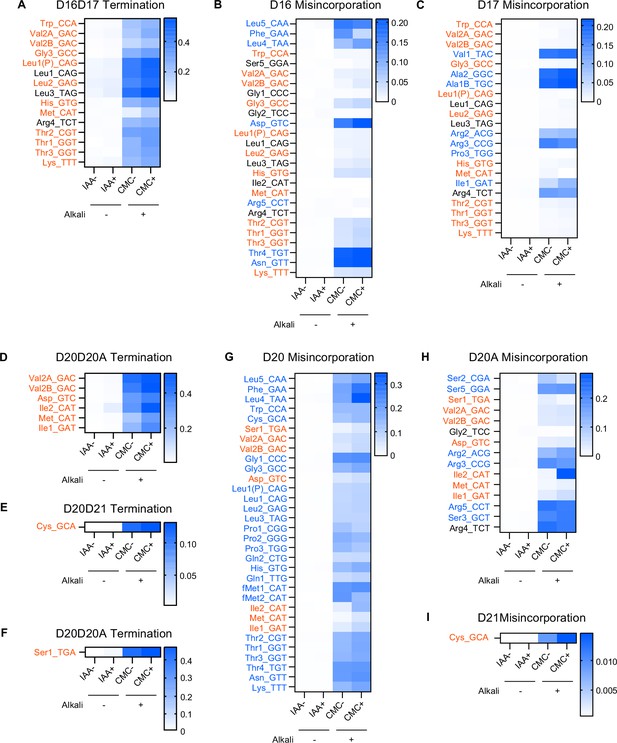
Misincorporation and termination signals derived from D in E. coli tRNAs.
Heatmaps showing termination frequencies at positions 18 (A), 21 (D, E), and 22 (F), and misincorporation frequencies at positions 16 (B), 17 (C), 20 (G), 20A (H), and 21 (I) to predict the presence of D. tRNAs bearing U at the indicated positions are shown. tRNA species bearing two consecutive D at positions are shown in red, whereas tRNA species containing single D at positions are shown in blue. D at a single position appears to induce misincorporation, and consecutive D likely induces termination of reverse transcription at the following position. The RT signatures are elevated in CMC− and CMC+ conditions, which include alkali treatment. Strain and chemical treatment are shown at the bottom.
-
Figure 5—figure supplement 3—source data 1
Termination frequencies at positions 18, 21, and 22, and misincorporation frequencies at positions 16, 17, 20, 20A, and 21 to predict the presence of D in E. coli tRNAs.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig5-figsupp3-data1-v1.xlsx
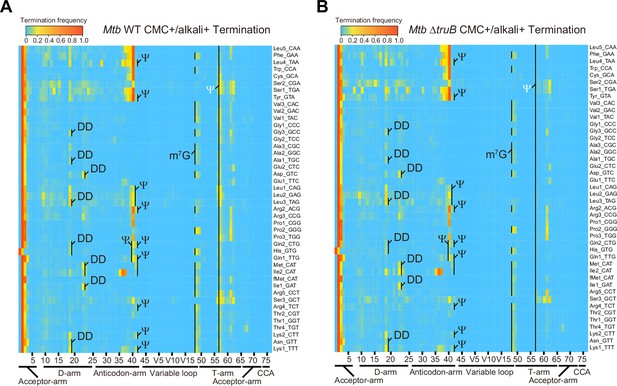
Heatmap of early termination frequency from sequencing of tRNAs isolated from wild-type (WT) and MtbΔtruB following 1-cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) treatment.
Heatmap of early termination frequencies across tRNA molecules and positions for WT (left) and MtbΔtruB (right). Termination signals derived from position 55 are shown in white. The experiment was performed once.
-
Figure 6—source data 1
Termination frequencies across tRNA molecules and positions for wild-type (WT) and MtbΔtruB.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig6-data1-v1.xlsx
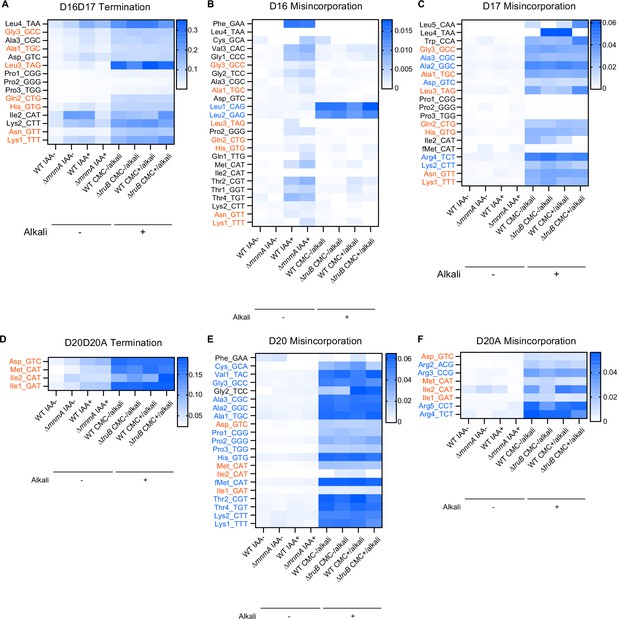
Misincorporation and termination signals derived from D at positions 20, 20A, and 21 in Mtb tRNAs.
Heatmaps showing termination frequencies at positions 18 (A) and 21 (D), and misincorporation frequencies at positions 16 (B), 17 (C), 20 (E), and 20A (F) to predict the presence of D. tRNAs bearing U at the indicated positions are shown. tRNA species bearing two consecutive D are shown in red, whereas tRNA species containing single D are shown in blue. D at a single position appears to induce misincorporation, and consecutive D likely induces termination of reverse transcription at the following position. The RT-signatures are elevated in CMC− and CMC+ conditions, which include alkali treatment. Strain and chemical treatment are shown at the bottom.
-
Figure 6—figure supplement 1—source data 1
Termination frequencies at positions 18 and 21, and misincorporation frequencies at positions 16 (B), 17, 20, and 20A to predict the presence of D in Mtb tRNAs.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig6-figsupp1-data1-v1.xlsx
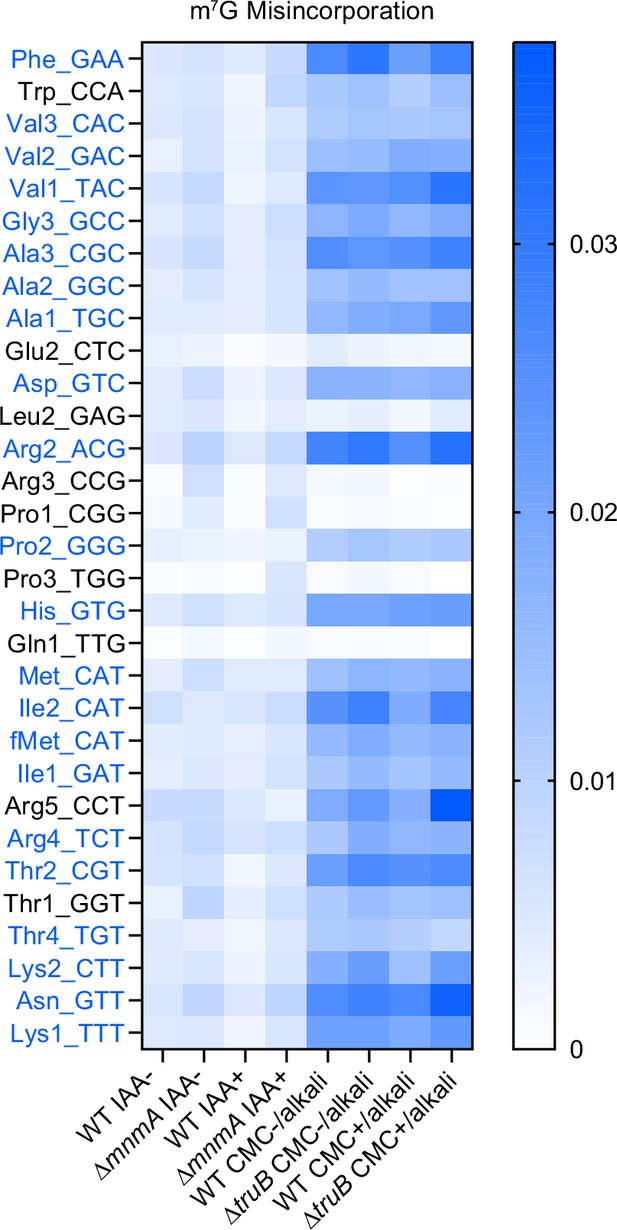
Misincorporation signals derived from m7G at position 46.
Heatmaps showing termination frequencies at position 46 to predict the presence of m7G. tRNAs bearing G at position 46 are shown. tRNAs that have consistently high signals when tRNAs are treated alkali are shown in blue. Strain and chemical treatment are shown in bottom.
-
Figure 6—figure supplement 2—source data 1
Termination frequencies at position 46 to predict the presence of m7G in Mtb tRNAs.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig6-figsupp2-data1-v1.xlsx
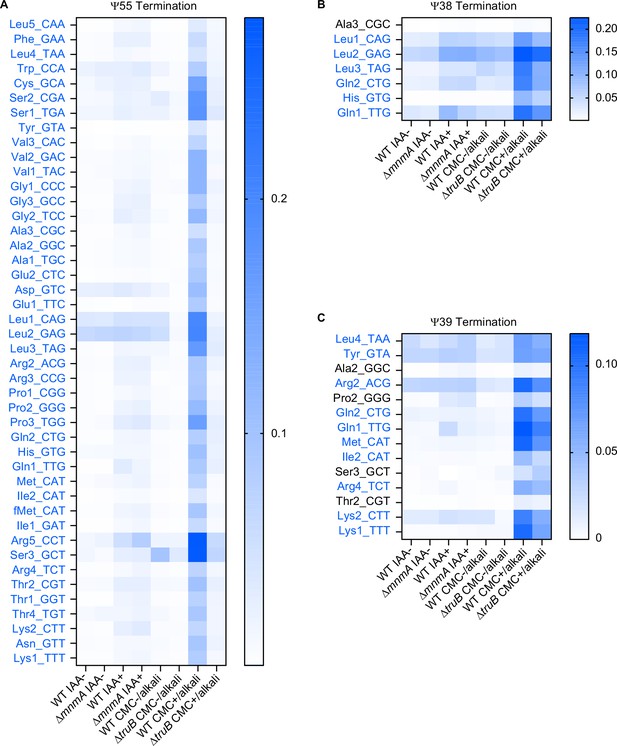
Termination signals derived from pseudouridine at positions 55, 38, and 39.
Heatmaps showing termination frequencies at positions 57 (A), 40 (B), and 41 (C) to assess the pseudouridylation states at positions 55 (A), 38 (B), and 39 (C). tRNAs bearing U at the indicated positions are shown and tRNAs that have higher signals when 1-cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) treated samples are shown in blue. Strain and chemical treatment are shown in bottom.
-
Figure 6—figure supplement 3—source data 1
Termination frequencies at positions 57, 40, and 41 to assess the pseudouridylation states at positions 55, 38, and 39 in Mtb tRNAs.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig6-figsupp3-data1-v1.xlsx
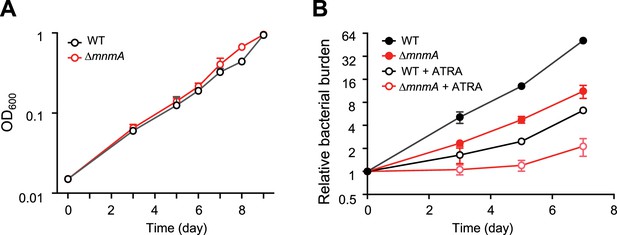
MtbΔmnmA is attenuated in a macrophage infection model.
(A) Wild-type and MtbΔmnmA do not display growth differences in 7H9 medium. (B) Auto-luminescent wild-type and ΔmnmA Mtb strains were diluted to a multiplicity of infection of 2 bacteria per mouse bone marrow-derived macrophage with or without all-trans retinoic acid (ATRA). Survival was measured by luminescence and normalized to luminescence at time 0. Average values from three independent cultures (n=3) are shown with standard deviations.
-
Figure 7—source data 1
Mtb growth curve.
- https://cdn.elifesciences.org/articles/87146/elife-87146-fig7-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mycobacterium tuberculosis) | mnmA | NA | Uniprot: Rv3024c; Refseq: NP_217540.1 | |
| Gene (Mycobacterium tuberculosis) | truB | NA | Uniprot: Rv2793c; Refseq: NP_217309.1 | |
| Strain, strain background (Mycobacterium tuberculosis) | H37Rv | PMID:9634230 | ||
| Genetic reagent (Mycobacterium tuberculosis) | MtbΔmnmA::zeo | This paper | Mtb H37Rv strain lacking mnmA | |
| Genetic reagent (Mycobacterium tuberculosis) | MtbΔtruB::zeo | This paper | Mtb H37Rv strain lacking truB | |
| Recombinant DNA reagent | pNit-RecET SacBR | NA | For homologous recombination | |
| Chemical compound | Iodoacetamide (IAA) | Sigma | ||
| Chemical compound | 1-Cyclohexyl-(2-morpholinoethyl) carbodiimide (CMC) | Sigma |
Additional files
-
Supplementary file 1
tRNA modifying enzymes used as queries for BLAST.
- https://cdn.elifesciences.org/articles/87146/elife-87146-supp1-v1.xlsx
-
Supplementary file 2
Exploration of tRNA modifying enzymes in Mtb by BLAST.
- https://cdn.elifesciences.org/articles/87146/elife-87146-supp2-v1.xlsx
-
Supplementary file 3
Predicted tRNA modifying enzymes in Mtb.
- https://cdn.elifesciences.org/articles/87146/elife-87146-supp3-v1.xlsx
-
Supplementary file 4
List of primers, plasmids, and strains.
- https://cdn.elifesciences.org/articles/87146/elife-87146-supp4-v1.xlsx
-
Supplementary file 5
Mtb tRNA sequences.
- https://cdn.elifesciences.org/articles/87146/elife-87146-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87146/elife-87146-mdarchecklist1-v1.pdf