Light-inducible protein degradation in E. coli with the LOVdeg tag
Figures

Design of AsLOV2-based degradation tag.
(a) Primary sequence of AsLOV2(546) C-terminal sequence. A three amino acid truncation exposes E-A-A. (b) Structure of AsLOV2 (aa404-546, PDB: 2V1A). Amino acids 541–543 (E-A-A) are red and 544–546 (K-E-L) are gray at the C-terminal of the Jα helix. (c) Construct used to characterize optogenetic control using AsLOV2 variants. Each variant is translationally fused to mCherry expressed from an IPTG-inducible promoter. Variants include wild-type AsLOV2 (light blue) and a dark state-stabilized version, AsLOV2* (dark blue), with and without the three amino acid truncation. (d) mCherry protein levels in response to 465 nm blue light for wild-type AsLOV2, and mutated AsLOV2* fusions with and without truncation. AsLOV2*(543) is the variant we denote the ‘LOVdeg’ tag. (***p<0.0001; **p<0.001; *p<0.01; n.s., not significant; two-tailed unpaired t-test; n = 3 biological replicates). (e) mCherry-LOVdeg in response to variable light intensities. (f) mCherry fluorescence levels and optical density of mCherry-LOVdeg with 4 hr of 465 nm blue light exposure applied at different points in the growth cycle. Light exposure programs are plotted above each subplot and are staggered 2 hr apart (starting at 2, 4, 6, or 8 hr), all lasting 4 hr. Expression levels are normalized to the dark state control (Figure 1—figure supplement 8). Error bars show standard deviation around the mean.

Alignment of AsLOV2, iLID (mutated version of AsLOV2 from Guntas et al., 2015) and LOVdeg (AsLOV2*(543)) amino acid sequences.
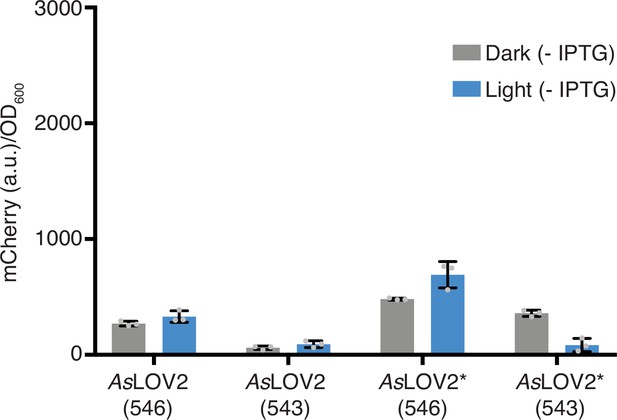
mCherry expression levels without IPTG induction in response to 465 nm blue light for wild-type AsLOV2 and mutated AsLOV2* fusions with and without truncation.
Error bars show standard deviation around the mean (n = 3 biological replicates).
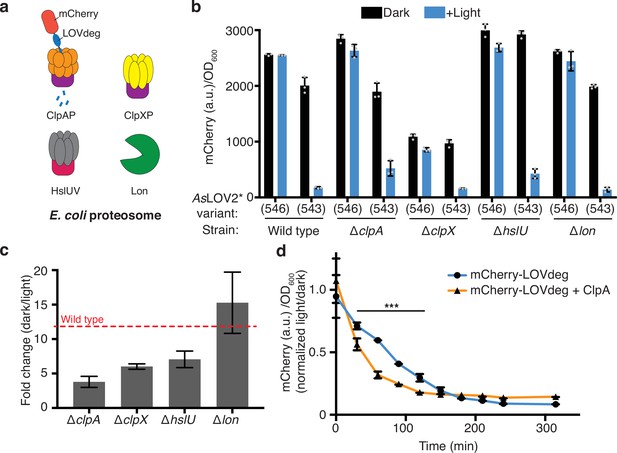
Investigating proteasome components involved in LOVdeg tag destabilization.
(a) Unfoldases and proteases of the E. coli proteasome. (b) Light-dependent stability of constitutively expressed mCherry fusions with truncated (AsLOV2*(543), LOVdeg) and non-truncated (AsLOV2*(546)) tags in strains lacking endogenous unfoldases or proteases. (c) Fold change degradation of mCherry-LOVdeg (i.e., mCherry-AsLOV2*(543)) in strains lacking endogenous unfoldases. Fold change compares ratio of dark to light states. (d) Expression of mCherry-LOVdeg over time under light exposure in wild-type cells or cells overexpressing ClpA. Fluorescence signal is normalized to expression of cells kept in the dark (***p<0.0001, comparison between data at the same time point, two-tailed unpaired t-test). Error bars show standard deviation around the mean (n = 3 biological replicates).
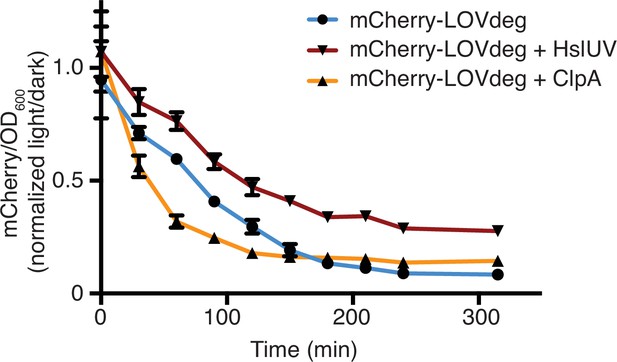
Expression of mCherry-LOVdeg over time under light exposure in wild-type cells, cells expressing exogenous HslUV, and cells expressing exogenous ClpA.
Fluorescence signal is normalized to expression of cells kept in the dark. Error bars show standard deviation around the mean (n = 3 biological replicates). ClpA expression data is the same as that shown in Figure 1—figure supplement 3 and included for comparison.
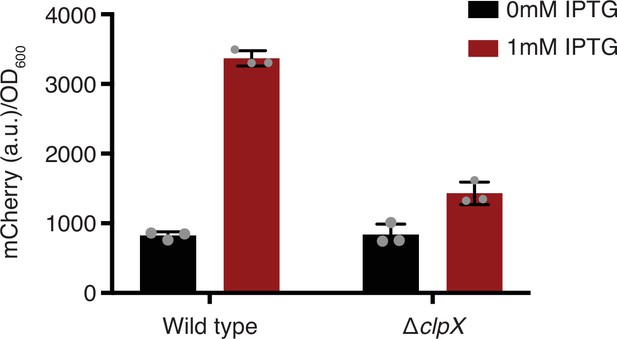
Untagged mCherry expression induced with IPTG in wild-type and clpX knockout strains.
Error bars show standard deviation around the mean (n = 3 biological replicates).
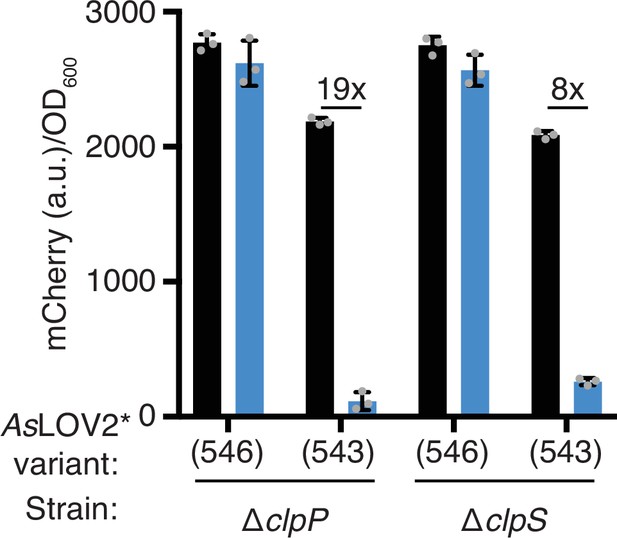
Light-dependent stability of mCherry fusions with truncated and non-truncated LOVdeg tags in strains lacking clpP and clpS.
Error bars show standard deviation around the mean (n = 3 biological replicates).
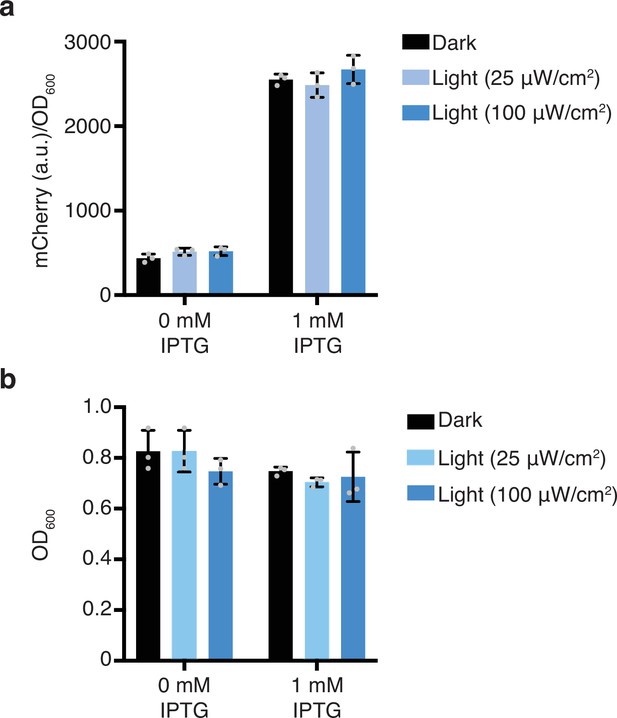
Light response of mCherry without any AsLOV2 variant.
(a) mCherry protein levels in response to dark or 465 nm blue light. (b) Culture densities (OD600) of strains from (a). Error bars show standard deviation around the mean (n = 3 biological replicates).
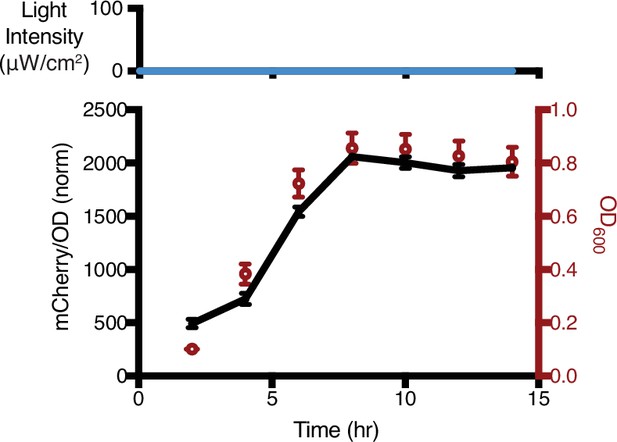
mCherry-LOVdeg fluorescence levels and growth without any light exposure.
This curve is used to normalize data in Figure 1f at each time point so that it is possible to see relative decreases in protein levels. Error bars show standard deviation around the mean (n = 3 biological replicates).
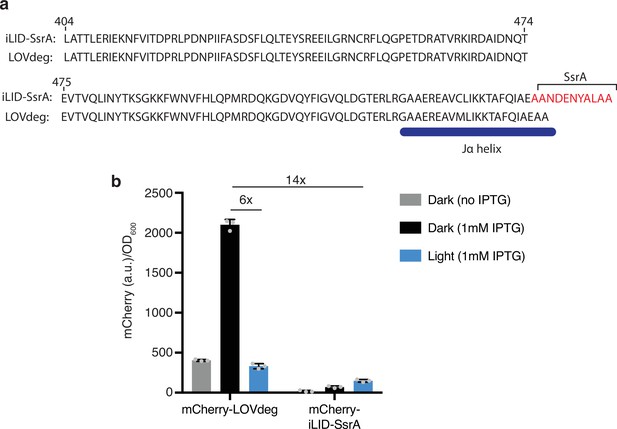
Comparing LOVdeg and iLID-SsrA.
(a) Alignment of iLID modified to contain a full-length SsrA tag and the LOVdeg tag. (b) Protein-level comparison between mCherry-LOVdeg and an analog with a constitutively active SsrA tag. Error bars show standard deviation around the mean (n = 3 biological replicates).
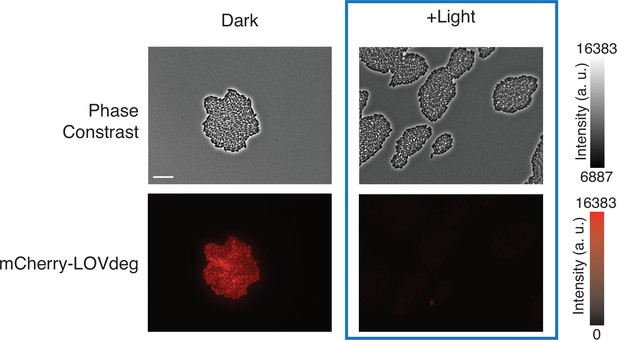
Phase contrast and fluorescence images of cells constitutively expressing mCherry-LOVdeg exposed to blue light or kept in the dark (scale bar = 10 μm).
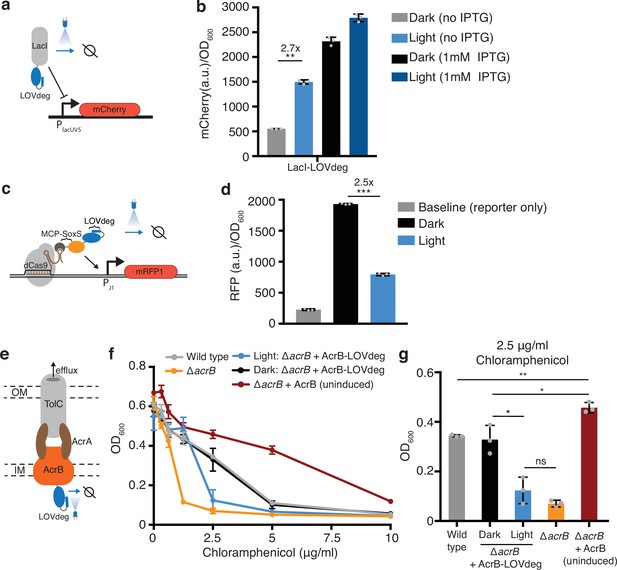
Incorporating light responsiveness into diverse proteins with the LOVdeg tag.
(a) Control of mCherry repression using a LacI-LOVdeg fusion. (b) mCherry expression in response to light exposure for strains with LacI-LOVdeg compared to IPTG induction (**p<0.001, two-tailed unpaired t-test). (c) Schematic of SoxS-based CRISPRa activation with a LOVdeg tag appended to the MCP-SoxS activator. (d) CRISPRa control of mRFP1 expression in response to light (***p<0.0001, two-tailed unpaired t-test). (e) Schematic of the LOVdeg tag appended to AcrB of the AcrAB-TolC efflux pump. IM, inner membrane; OM, outer membrane. (f) Chloramphenicol sensitivity tests. Wild-type cells (BW25113) are compared to a ΔacrB (BW25113 ΔacrB) strain, ΔacrB complemented with AcrB-LOVdeg (ΔacrB + AcrB-LOVdeg) exposed to light or kept in the dark, and ΔacrB strain complemented with an IPTG-inducible AcrB (ΔacrB + AcrB). No IPTG was added to ΔacrB + AcrB or ΔacrB + AcrB-LOVdeg. (g) OD600 of strains shown in (f) at 2.5 μg/mL chloramphenicol (**p<0.001; *p<0.05; ns, not significant; two-tailed unpaired t-test). Error bars show standard deviation around the mean (n = 3 biological replicates).
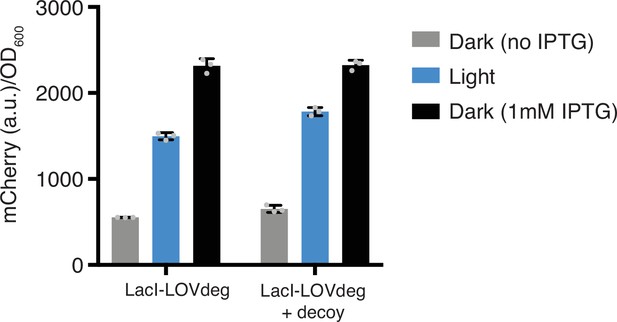
LacI-LOVdeg and LacI-LOVdeg+decoy control of mCherry with 1 mM IPTG induction included.
Error bars show standard deviation around the mean (n = 3 biological replicates).
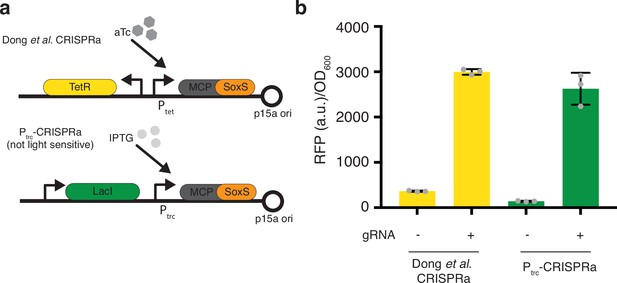
Switching promoters for the MCP-SoxS construct.
(a) Genetic constructs of the original SoxS-CRISPRa from Dong et al., where the activator is anhydrotetracycline (aTc) inducible. However, aTc is light sensitive, thus we replaced the TetR/Ptet portion with the IPTG-inducible LacI/Ptrc. (b) Comparison of the CRISPRa activity of the original construct and the new Ptrc promoter construct with and without a gRNA targeting the promoter of mRFP1. Error bars show standard deviation around the mean (n = 3 biological replicates).
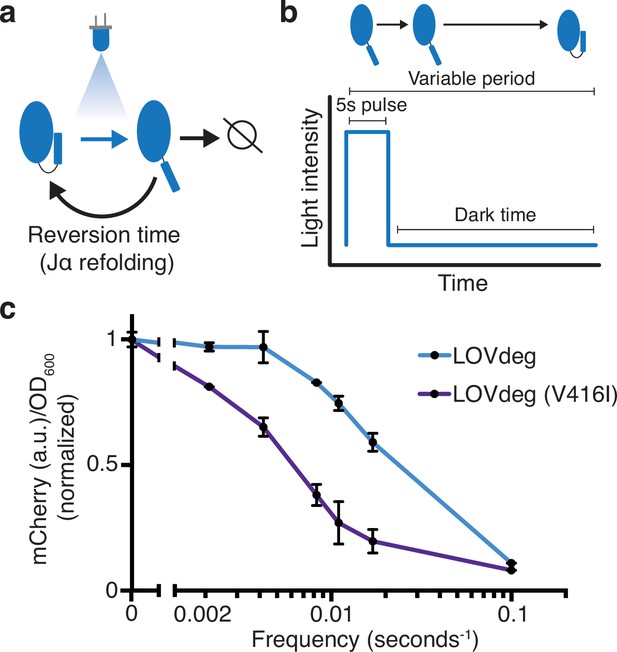
Modulating LOVdeg frequency response with photocycle mutations.
(a) Photocycle of AsLOV2. Upon light absorption, the Jα helix unfolds for a period of time dictated by the stability of the light state conformation. If not degraded, the Jα helix refolds, blocking degradation. (b) The light program used to test frequency responses of LOVdeg photocycle variants in (c). A constant pulse of 5 s is followed by a variable dark time that allows for Jα helix refolding. (c) Expression of mCherry-LOVdeg and variant mCherry-LOVdeg (V416I) in response to different light exposure frequencies. Fluorescence values are normalized to dark state expression. Error bars show standard deviation around the mean (n = 3 biological replicates).

Enhanced light response using EL222 transcriptional control together with LOVdeg.
(a) mCherry-LOVdeg expressed from an EL222-responsive promoter that is constitutively active in the absence of EL222. Addition of EL222 represses mCherry production. These two forms of regulation are combined when mCherry-LOVdeg is expressed from an EL222-responsive promoter, resulting in a circuit that both degrades and represses in response to light. (b) Light and dark expression of mCherry in the ‘degradation only’ (closed circles), ‘repression only’ (open circles), or ‘repression + degradation’ (squares) strains. Error bars show standard deviation around the mean (n = 3 biological replicates).
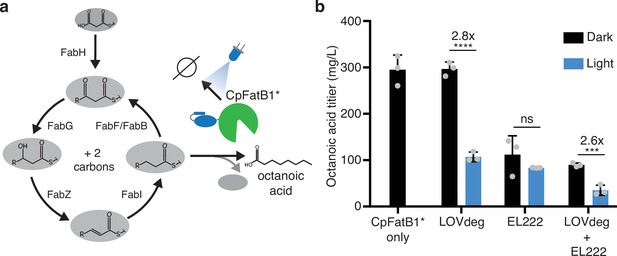
Optogenetic control of octanoic acid production.
(a) Schematic of fatty acid synthesis in E. coli. CpFatB1* catalyzes elongating C8-ACP molecules from this pathway to produce free octanoic acid. CpFatB1* is tagged with LOVdeg to create optogenetic control. (b) Octanoic acid titer from strains that express CpFatB1* only, CpFatB1*-LOVdeg only, EL222 regulated CpFatB1* only, or CpFatB1*-LOVdeg+EL222. Octanoic acid is quantified by GC-MS. Strains were kept either in the dark or with continuous blue light exposure for the duration of the production period. Error bars show standard deviation around the mean (****p<0.0001; ***p<0.0001; ns, not significant; two-tailed unpaired t-test; n = 3 biological replicates).
Additional files
-
Supplementary file 1
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/87303/elife-87303-supp1-v1.xlsx
-
Supplementary file 2
Strains used in this study.
- https://cdn.elifesciences.org/articles/87303/elife-87303-supp2-v1.xlsx
-
Supplementary file 3
DNA sequences used in this study.
- https://cdn.elifesciences.org/articles/87303/elife-87303-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87303/elife-87303-mdarchecklist1-v1.docx





