A large-scale proteomics resource of circulating extracellular vesicles for biomarker discovery in pancreatic cancer
Figures
Study design.
The discovery cohort was comprised of 124 individuals, including pancreatic ductal adenocarcinoma (PDAC, N=93), chronic pancreatitis (CP, N=12), intraductal papillary mucinous neoplasm (IPMN, N=8) and healthy controls (N=11). Plasma samples were processed for EV isolation using EVtrap and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
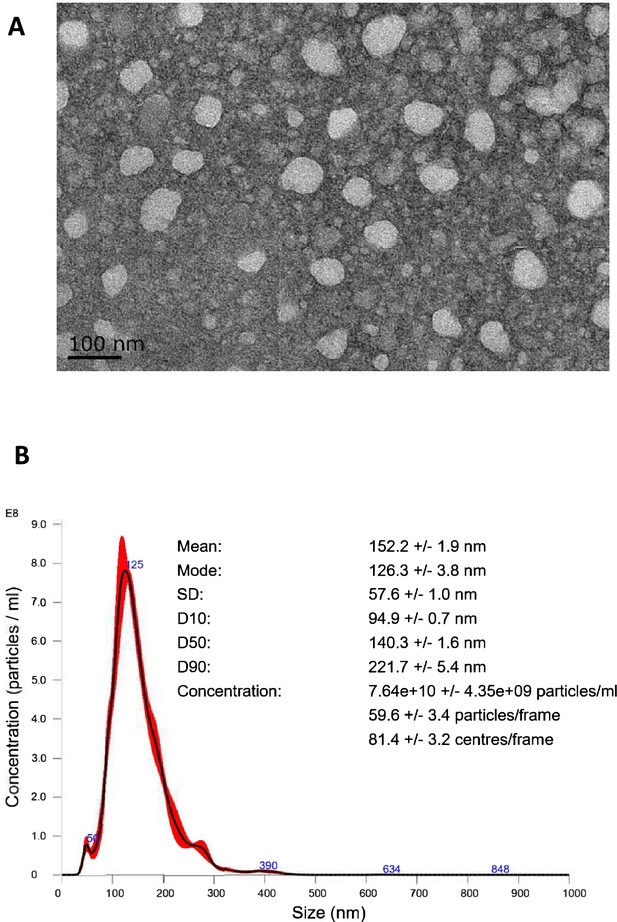
EVtrap isolation of extracellular vesicles.
(A) Transmission electron microscopy (TEM) images collected of a single EV and multiple EVs captured from plasma by EVtrap. TEM imaging of EVs was carried out on a HITACHI H-8100 electron microscope (Hitachi, Tokyo, Japan) with an accelerating applied potential of 200 kV. (B) Nanoparticle tracking analysis (NTA) of EVs after elution off EVtrap beads. NTA was carried out using ZetaView instrument (Particle Metrix) after calibration with 100 nm polystyrene particles.
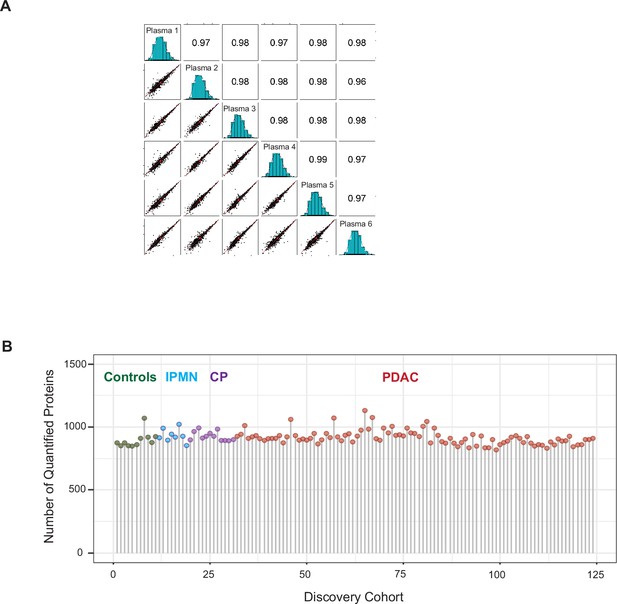
EV proteomics analytical performance.
(A) Reproducibility of the method. A standard plasma sample was processed in six replicates and performed a Pearson correlation analysis that revealed a very high correlation between replicates. (B) Number of quantified EV proteins per sample according to different patient cohort.
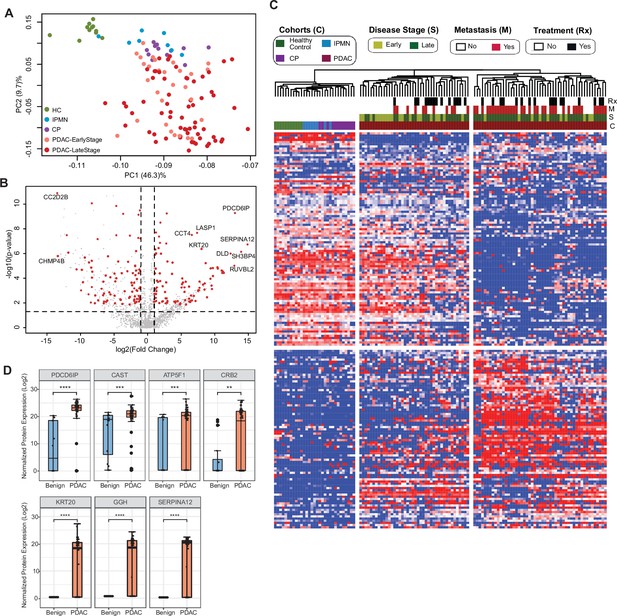
Identification of cEV proteins differentially expressed in disease groups.
(A) Principal component analysis of cEV proteins differentially expressed in the plasma of patients with pancreatic diseases compared to controls. Each dot indicates one individual enrolled in the study: green, controls; blue, patients with intraductal papillary mucinous neoplasm (IPMN); purple, patients with chronic pancreatitis (CP); salmon, early stage (stages I and II) pancreatic ductal adenocarcinoma (PDAC); red, late stage (stages III and IV) PDAC. (B) Volcano plot of circulating EV proteins enriched in the plasma of patients with PDAC versus benign pancreatic diseases. X-axis, log base 2 of fold changes; Y-axis, negative of the log base 10 of p values. (C) Heatmap of cEV proteins differentially expressed in the plasma of patients with pancreatic diseases compared to controls. Designations of clinical parameters were indicated at the top of the heatmap. (D) Expression of enriched cEV proteins in patients with PDAC (N=93) versus benign pancreatic diseases (N=20). Each dot indicates the target protein signal from one patient. Y-axis, normalized log base 2 of protein signals detected by mass spectrometry; Error bars, min and max values; lines in boxes, median values. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
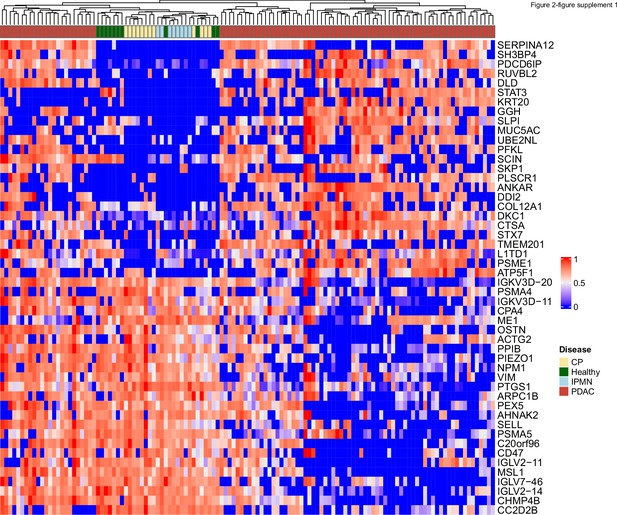
Heatmap of abundance of 25 proteins enriched and 25 proteins reduced in EVs from PDAC patients compared to EVs from patients without cancer.
Protein abundances were normalized across patients for each protein.
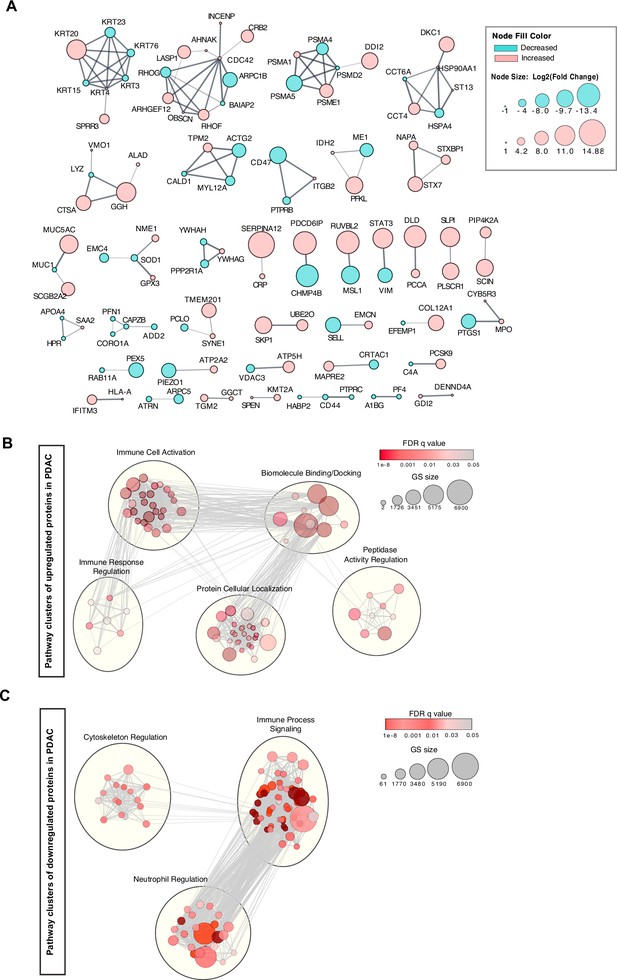
Network analyses of cEV proteins differentially expressed in PDAC compared to benign pancreatic diseases.
(A) Functional association of proteins identified by STRING database. Red, cEV proteins enriched in PDAC patients as compared to benign pancreatic diseases. Green, cEV proteins decreased in patients with PDAC as compared to benign pancreatic diseases. Red, cEV proteins increased in PDAC as compared to benign pancreatic diseases. Thickness of lines indicate confidence of association. (B, C) Clustering of cEV protein pathways enriched (B) or downregulated (C) in PDAC cohorts. Pathways were identified using Gene Ontology database and REACTOME database.
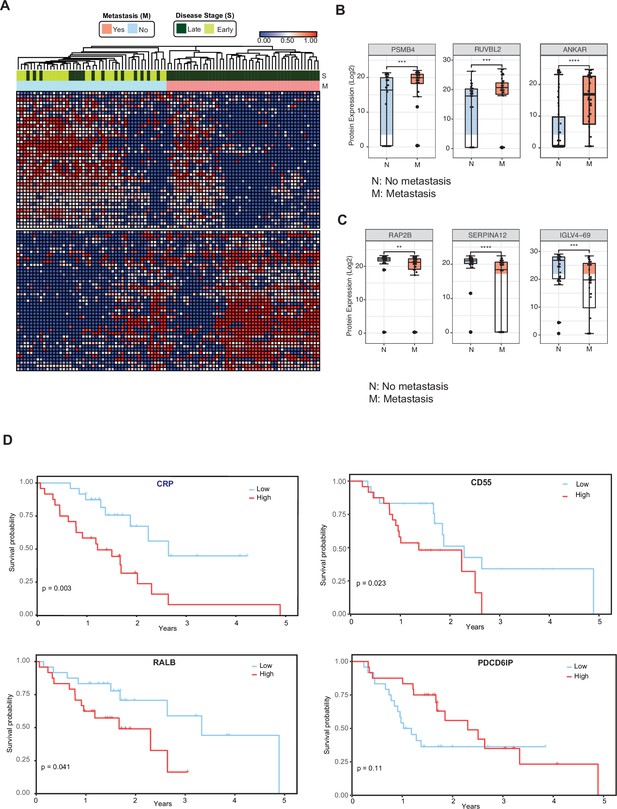
Circulating EV proteomics reveal markers associated with metastasis and worse prognosis.
(A) Heatmap showing EV proteins differentially expressed in the plasma of metastatic versus non-metastatic PDAC. Designations of clinical parameters are indicated at the top of the heatmap. (B) Expression patterns of cEV proteins associated with metastatic disease. Y-axis, normalized log base 2 of protein signals detected by mass spectrometry; N, non-metastatic PDAC group (N=46); M, metastatic PDAC group (N=47). Each dot indicated the target protein signal from one patient. Error bars, min and max values; lines in boxes, median values. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001. (C) As is (B), except for cEV markers with increased expression in non-metastatic PDAC. (D) Correlation of cEV marker expression with survival. Kaplan–Meier curves and log-rank test p values of representative survival cEV markers quantified in the discovery cohort.
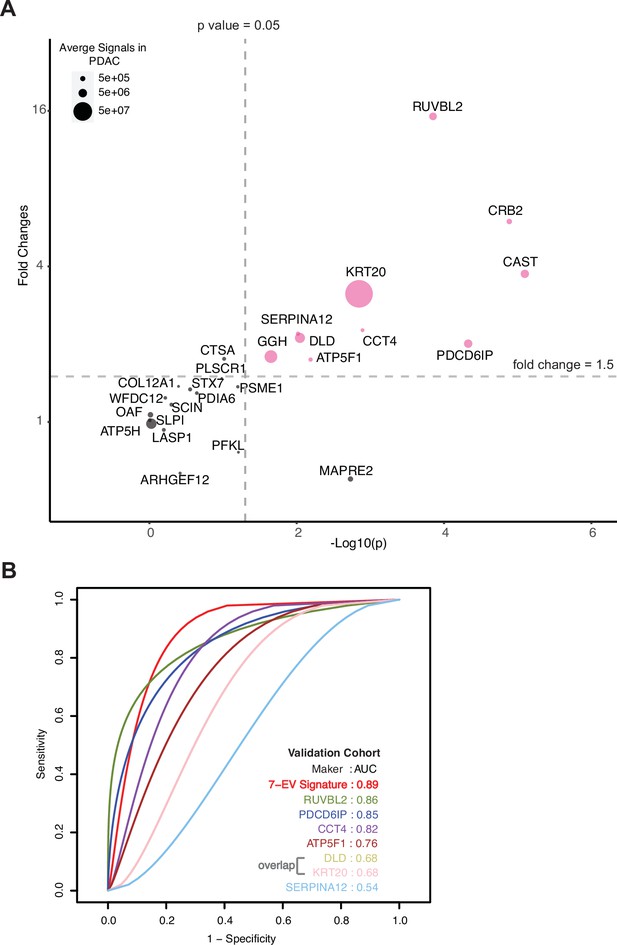
Validation of cEV markers and identification of seven EV protein signature for PDAC diagnosis.
(A) Differences of cEV protein abundances between patients with PDAC (n=24) and benign pancreatic diseases (chronic pancreatitis and IPMN) (n=12). x axis, minus log p values of protein abundance differences between PDAC and benign groups; y axis, average fold changes of proteins in PDAC group compared to benign group. Size of bubbles indicate average protein abundances in PDAC group. Pink color, proteins that had at least twofold enrichment in PDAC group (p<0.05). (B) ROC curves were calculated for individual cEV markers as well as for the seven EV protein PDAC signature combination to determine optimum diagnostic performance.
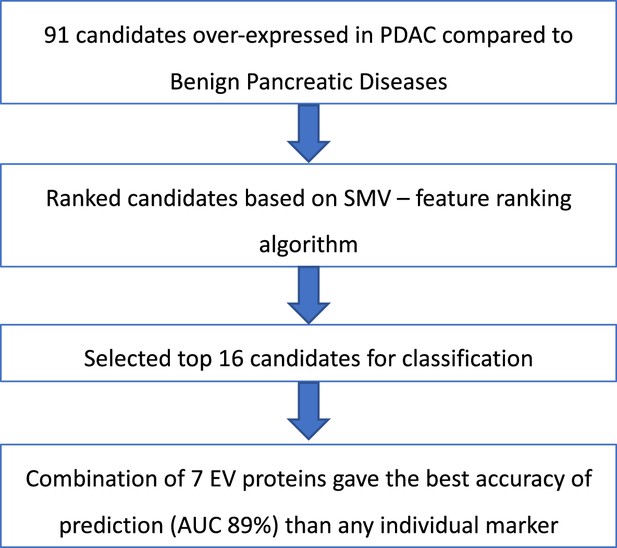
Summary of selection process to develop EV signature for pancreatic cancer diagnosis.
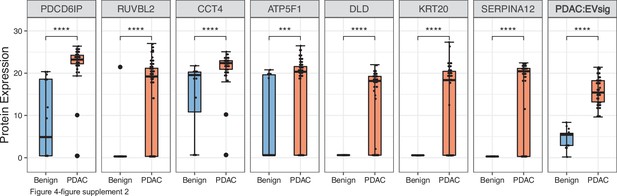
Diagnostic performance of the seven EV protein signature compared to performance of each of the seven individual marker in patients with benign pancreatic diseases (N=12) or PDAC (N=24).
Each dot indicated the target protein signal from one patient. Error bars, min and max values; lines in boxes, median values. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
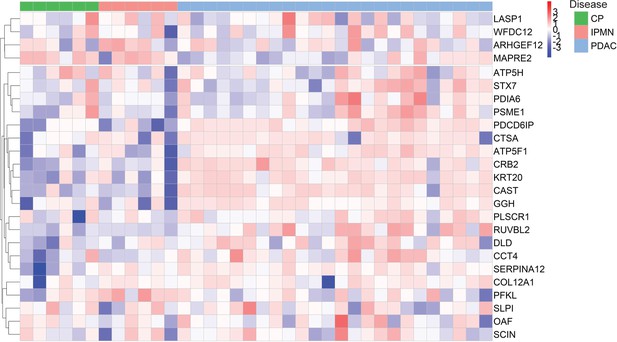
Validation of individual cEV proteins in an independent cohort of patients.
Expression of biomarker candidates detected by Parallel Reaction Monitoring (PRM) analyses. A total of 25 cEV proteins with significant overexpression in PDAC in the discovery cohorts were quantified by PRM in a separate validation cohort of patients.
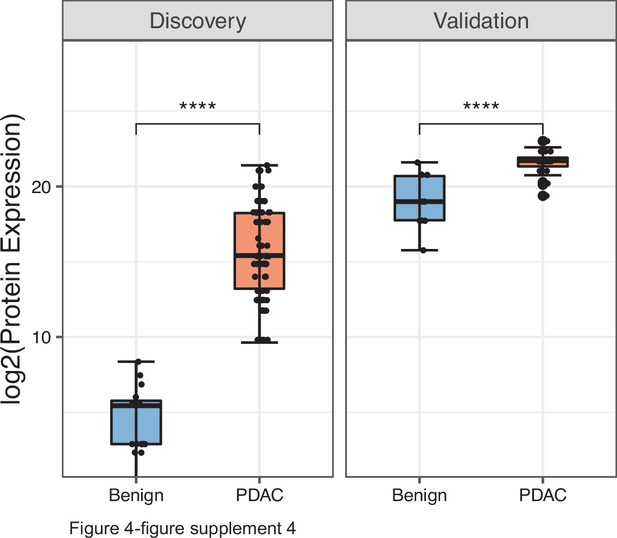
Performance of PDAC EV signature in both discovery (benign =20, PDAC=93) and validation (benign =12, PDAC=24) cohorts.
Each dot indicated the target protein signal from one patient. Error bars, min and max values; lines in boxes, median values. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
Additional files
-
Supplementary file 1
Baseline characteristics of patients enrolled on the discovery cohort.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp1-v1.xlsx
-
Supplementary file 2
Plasma EV analysis reproducibility.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp2-v1.xlsx
-
Supplementary file 3
LC-MS results of EV analysis of plasma from patients with PDAC (PA), IPMN, Chronic Pancreatitis (CP) and Control individuals.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp3-v1.xlsx
-
Supplementary file 4
List of EV proteins that met the eligibility criteria for principal component analysis.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp4-v1.xlsx
-
Supplementary file 5
List of 182 proteins differentially expressed in PDAC compared to benign diseases.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp5-v1.xlsx
-
Supplementary file 6
List of EV proteins that are significantly altered in patients with metastatic versus non-metastatic diseases.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp6-v1.xlsx
-
Supplementary file 7
Table A: Support Vector Machine Prediction model output for the 16 individual markers included in the in External Validation Cohorts.
Table B: The contingency table for 7-biomarker signature, offering insights into model accuracy for both the Internal-Discovery and External Validation cohorts.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp7-v1.xlsx
-
Supplementary file 8
List of 25 cEV proteins that met the eligibility criteria for validation studies.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp8-v1.xlsx
-
Supplementary file 9
Baseline characteristics of patients enrolled in the validation cohort.
- https://cdn.elifesciences.org/articles/87369/elife-87369-supp9-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87369/elife-87369-mdarchecklist1-v1.pdf





