Oral supplementation of gut microbial metabolite indole-3-acetate alleviates diet-induced steatosis and inflammation in mice
Figures
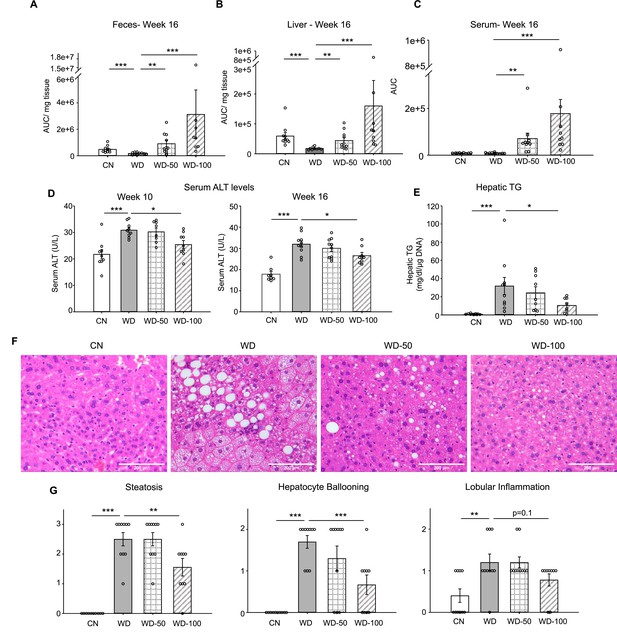
Oral administration of indole-3-acetate (I3A) alleviates diet-induced hepatic steatosis and inflammation.
(A) Fecal, (B) liver, and (C) serum concentrations of I3A in male B6 129SF1/J mice in control low-fat diet (CN), Western diet (WD), WD with low-dose I3A (WD-50), and WD with high-dose I3A (WD-100) at week 16. (D) Serum alanine aminotransferase (ALT) levels in mice at weeks 10 and 16. (E) Liver triglyceride (TG) levels at week 16. Data shown are TG concentrations (mg/dl) normalized to corresponding tissue DNA contents (µg DNA). (F) Representative liver sections stained with hematoxylin-eosin (H&E). (G) Histology score for steatosis, hepatocyte ballooning, and lobular inflammation. H&E-stained liver sections were evaluated by an expert pathologist using the non-alcoholic steatohepatitis (NASH) CRN and fatty liver inhibition of progression (FLIP) consortia criteria. Data shown are mean ± SEM (n=10 per group). *: p<0.05, **: p<0.01, ***: p<0.001 using Wilcoxon rank sum test.
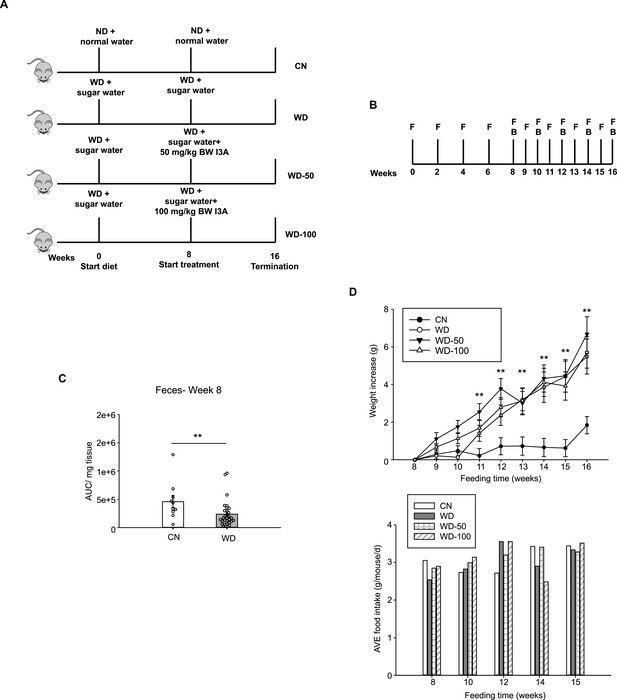
Overview of in vivo study.
(A) Three groups of male B6 129SF1/J mice (n=10 for each group) were fed ad libitum a Western diet (WD) and a sugar water (SW) solution while a fourth group was given normal chow diet. After 8 weeks, the three groups of WD-fed mice were randomly selected for treatment with vehicle (WD group) or low (WD-50 group) or high dose (WD-100 group) of I3A for an additional 8 weeks. The fourth group (control mice [CN]) was continued on low-fat diet calorie matched with the WD. (B) Sampling scheme. (C) Quantification of I3A in feces at week 8. (D) Weight increase and food intake of the four groups. Body weights were normalized to week 8 body weight when I3A administration was started. Food intakes were measured per cage and average food intake was calculated as per gram food intake per mouse per day. Data shown are mean ± SEM from a replicate study. **: p<0.01, WD group compared to CN group using Wilcoxon rank sum test.
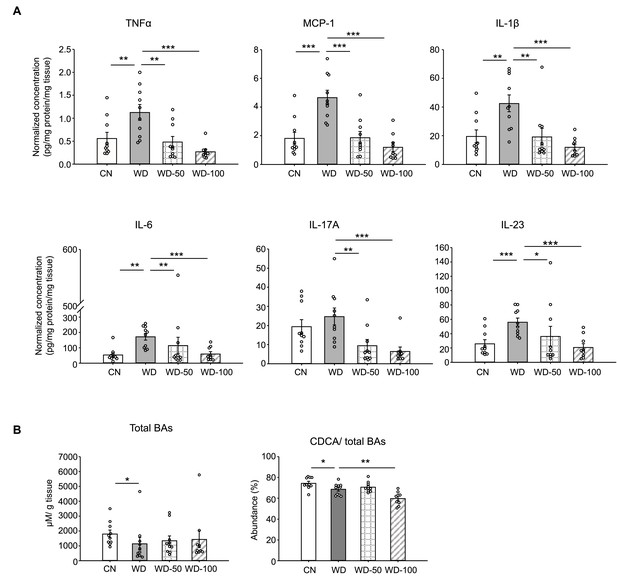
Indole-3-acetate (I3A) administration reverses Western diet (WD)-induced alterations in liver inflammatory cytokines and bile acids.
(A) Inflammatory cytokines in liver tissue at week 16. (B) Liver total bile acid concentration (left panel), and abundance of chenodeoxycholic acid (CDCA) branch bile acids relative to total bile acids pool (right panel). Data shown are mean ± SEM. *: p<0.05, **: p<0.01, ***: p<0.001 using Wilcoxon rank sum test.
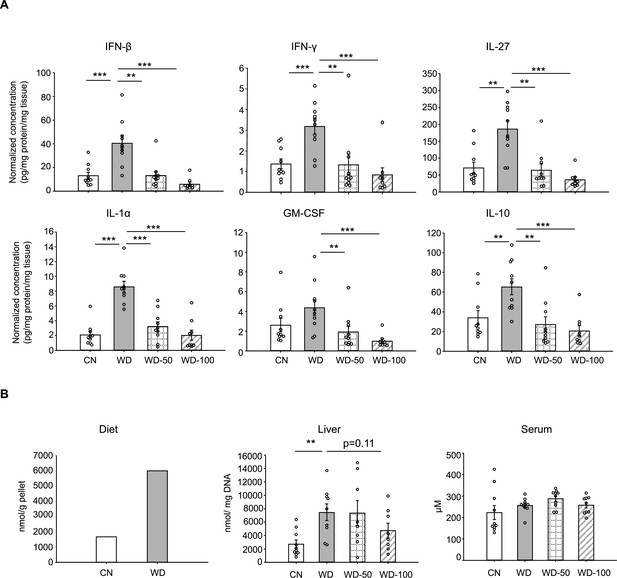
I3A supplementation reduces inflammatory cytokines and free fatty acid levels.
(A) Inflammatory cytokines in liver tissue at week 16. (B) Free fatty acids (FFAs) in diet, serum, and liver samples. Data shown are mean ± SEM. **: p<0.01, ***: p<0.001 using Wilcoxon rank sum test.
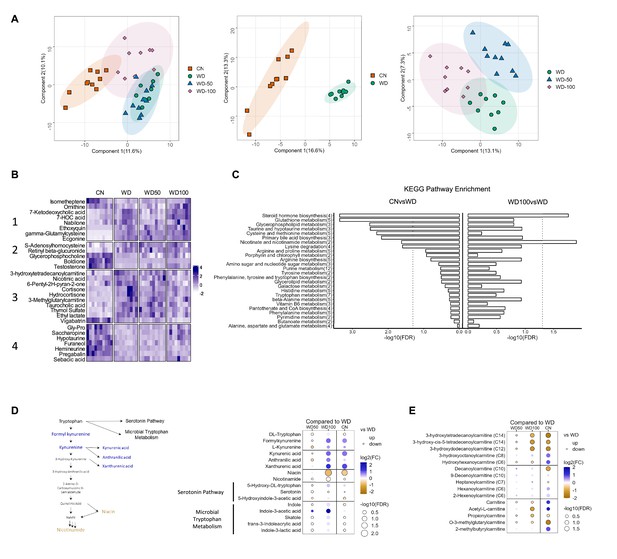
Indole-3-acetate (I3A) administration partially reverses diet-induced metabolome alterations in the liver.
(A) Scatter plots of latent variable projections from PLS-DA of untargeted metabolomics data features. Comparison of all four experimental groups (left panel), control mice (CN) vs. Western diet (WD) group (middle panel), and WD vs. WD-50 and WD-100 groups (right panel). (B) Heatmap of significant metabolite features (FDR<0.1) based on statistical comparisons of treatment groups (CN vs. WD). (C) KEGG pathway enrichment analysis of the metabolites. Number in the parenthesis represents the metabolites detected in the pathway. (D) Schematic for tryptophan metabolism (left panel). Tryptophan metabolism metabolites fold-changes of WD-50, WD-100, CN relative to WD (right panel). (E) Acyl-carnitine fold-change of WD-50, WD-100, CN relative to WD. p-Values were calculated using Student’s t-test and corrected by FDR.
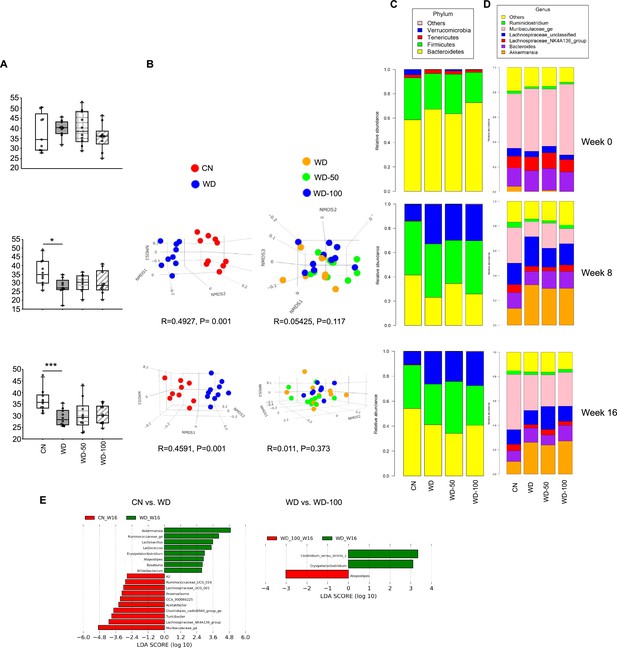
16srRNA metagenomics of mouse fecal microbiota.
(A) Alpha diversity of the fecal microbiome from control mice (CN), Western diet (WD), WD-50, and WD-100 groups. (B) Analysis of similarities (ANOSIM) comparison for CN vs. WD group and WD vs. WD-50 vs. WD-100 groups. (C, D) Phylum and genus level relative abundance of the fecal microbial community members. (E) Linear discriminant analysis effect size (LEfSe) results at the genus level. *: p<0.05, ***: p<0.001 using Wilcoxon rank sum test.
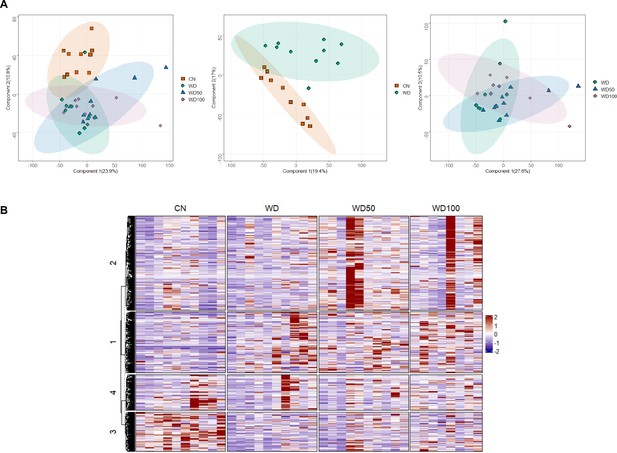
Untargeted metabolomic analysis of mouse fecal metabolites at week 16.
(A) Score plots show the first two principal components for all four experimental groups (left panel), control mice (CN) vs. Western diet (WD) group (middle panel), and WD vs. WD-50 and WD-100 groups (right panel). Numbers in the parentheses of axis titles show percent of variance explained by the corresponding principal component. Ellipses circumscribe 95% confidence regions for the experimental groups assuming Gaussian distribution of component scores. (B) Heatmap of fecal microbiome metabolite features detected in all treatment groups. Rows and columns are features and treatment groups, respectively. The features were clustered using k-means.
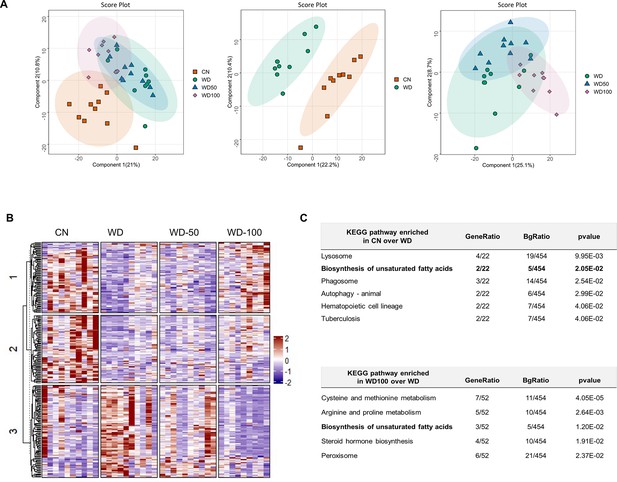
Indole-3-acetate (I3A) administration partially reverses diet-induced proteome alterations in the liver.
(A) Scatter plots of latent variable projections from PLS-DA of confidently identified proteins. Comparison of all four experimental groups (left panel), control mice (CN) vs Western diet (WD) group (middle panel), and WD vs. WD-50 and WD-100 groups (right panel). (B) Heatmap of significant proteins having variable importance in projection score >1.2. The proteins were clustered using k-means. (C) Pathway enrichment analysis of significant proteins differentially abundant in CN vs. WD comparison (upper panel) and WD-100 vs. WD comparison (lower panel). GeneRatio divides the number of significantly altered proteins that are in the pathway by the total number of significantly altered proteins. BGRatio divides the number of proteins that are in the pathway by the number of all detected proteins. The p-value was calculated using Fisher’s exact test.
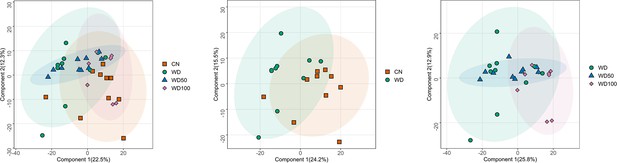
Score plots of the first two principal components for all four experimental groups (left panel), control mice (CN) vs. Western diet (WD) group (middle panel), and WD vs. WD-50 and WD-100 groups (right panel).
Numbers in the parentheses of axis titles show percent of variance explained by the corresponding principal component. Ellipses represent 95% confidence regions for the experimental groups assuming Gaussian distribution of component scores.
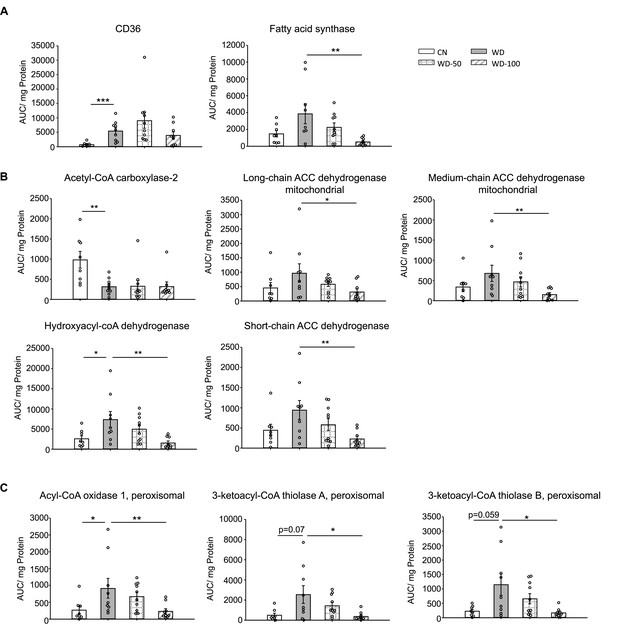
Indole-3-acetate (I3A) administration reduces the levels of enzymes in fatty acid transport, de novo lipogenesis, and β-oxidation.
(A) Abundance of fatty acid translocase (CD36) and fatty acid synthase. (B) Mitochondrial and (C) peroxisomal fatty acid oxidation enzymes. Data shown are mean ± SEM. *: p<0.05, **: p<0.01, ***: p<0.001 using Wilcoxon rank sum test.
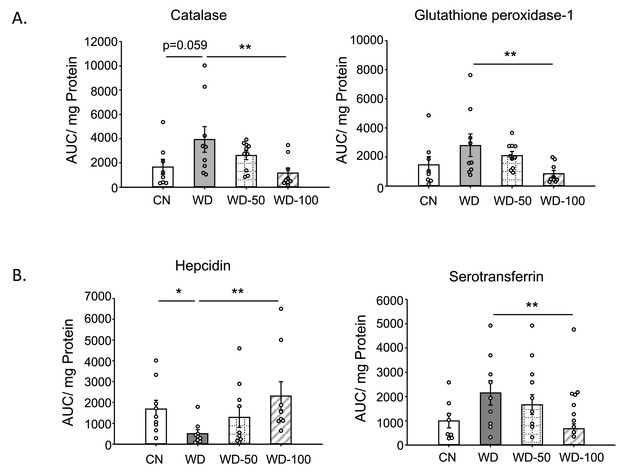
Targeted proteomic analysis of mouse liver tissue.
(A) Catalase and glutathione peroxidase-1 abundance for the four experimental groups. (B) Hepcidin and serotransferrin abundance. Data shown are mean ± SEM. *: p<0.05, **: p<0.01 using Wilcoxon rank sum test.
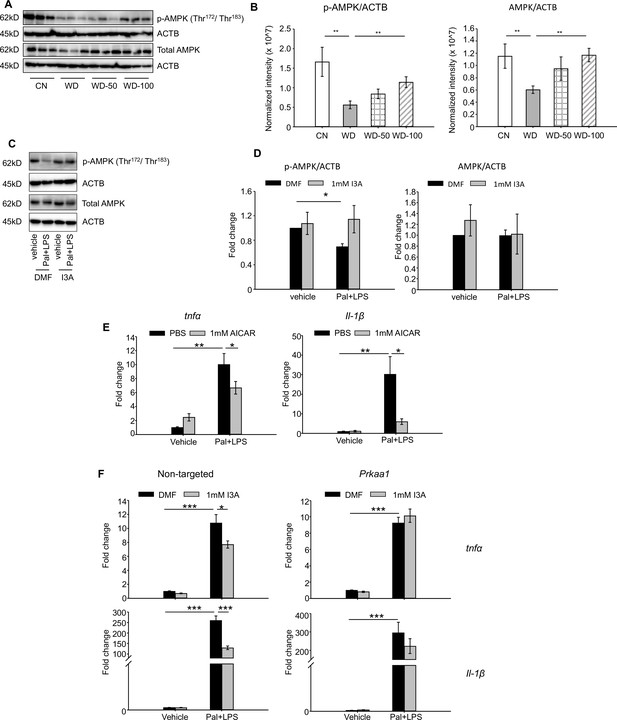
Indole-3-acetate (I3A) modulates AMP-activated protein kinase (AMPK) phosphorylation and suppresses RAW264.7 macrophage cell inflammation in an AMPK-dependent manner.
(A, B) I3A administration reverses Western diet (WD)-induced reduction in liver p-AMPK and AMPK. (A) Levels of p-AMPK and AMPK in liver tissue at week 16 as determined by western blot analysis. (B) Ratios of p-AMPK (left panel) and AMPK (right panel) to β-actin. The ratios were determined based on the p-AMPK and AMPK band intensities quantified using Image Lab (Bio-Rad) and normalized to the loading control (β-actin). Data shown are mean ± SEM. **: p<0.01 using Wilcoxon rank sum test. (C) Expression levels of p-AMPK and total AMPK in RAW 264.7 macrophages pre-treated with either I3A or vehicle (DMF) control followed by stimulation with palmitate and LPS, determined by western blot analysis. (D) Fold-changes in p-AMPK and total AMPK. Fold-changes were calculated relative to the DMF and no palmitate and LPS stimulation condition. The band intensities were quantified and normalized to loading control (β-actin) by using Image Lab (Bio-Rad). (E) Expression levels of Tnfa and Il1b in RAW 264.7 cells treated with p-AMPK activator 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), followed by stimulation with palmitate and LPS. (F) Expression levels of Tnfa (top row) and Il1b (bottom row) in RAW 264.7 cells transduced with non-targeted control siRNA (left panels) or Prkaa1 siRNA (middle panels), pre-treated with I3A, and then stimulated with palmitate and LPS. Data shown are mean ± SEM from three independent cultures with three biological replicates. *: p<0.05, **: p<0.01, ***: p<0.001 using Student’s t-test.
-
Figure 6—source data 1
Western blot analysis of p-AMPK and AMPK levels in mice liver tissues used in Figure 6A.
- https://cdn.elifesciences.org/articles/87458/elife-87458-fig6-data1-v1.pptx
-
Figure 6—source data 2
Western blot analysis of p-AMPK and AMPK levels in RAW264.7 cells used in Figure 6C.
- https://cdn.elifesciences.org/articles/87458/elife-87458-fig6-data2-v1.pptx
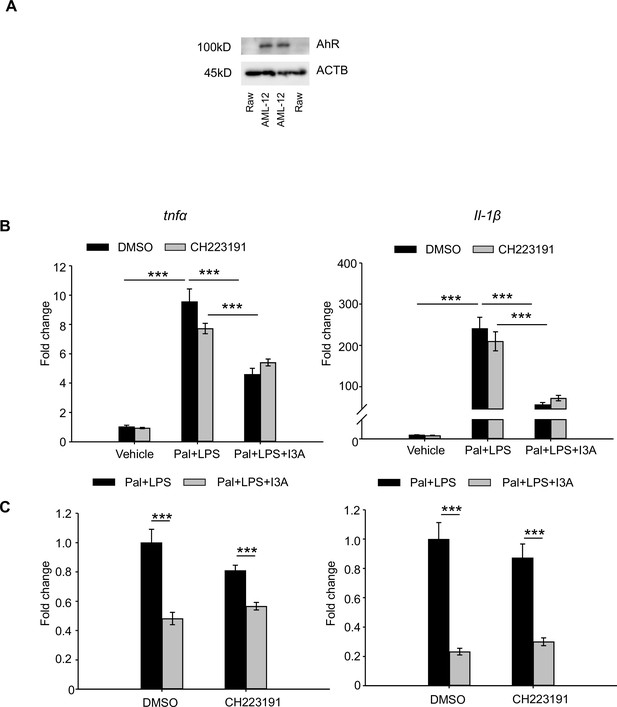
Anti-inflammatory effects of I3A on RAW264.7 cells is independent of AhR.
(A) Expression level of aryl-hydrocarbon receptor (AhR) in RAW264.7 and AML12 cells determined by western blot analysis. (B) Raw 264.7 cells were treated with 1 mM indole-3-acetate (I3A) (or DMF solvent control) for 4 hr, then stimulated with 300 µM palmitate for 18 hr and 10 ng/ml LPS for 6 hr (two-hit model). The AhR inhibitor CH223191 (5 µM) or DMSO control was added 10 min before I3A treatment. Total RNA was isolated from the cells and the expression of Tnfa and Il1b was measured with qRT-PCR. (C) Tnfa and Il1b expression was plotted as fold-change normalized to the DMF control group. Data presented as the mean ± SEM. ***: p<0.001 using Student’s t-test.
-
Figure 6—figure supplement 1—source data 1
Western blot analysis of AhR level in AML12 and RAW264.7 cells used in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/87458/elife-87458-fig6-figsupp1-data1-v1.pptx
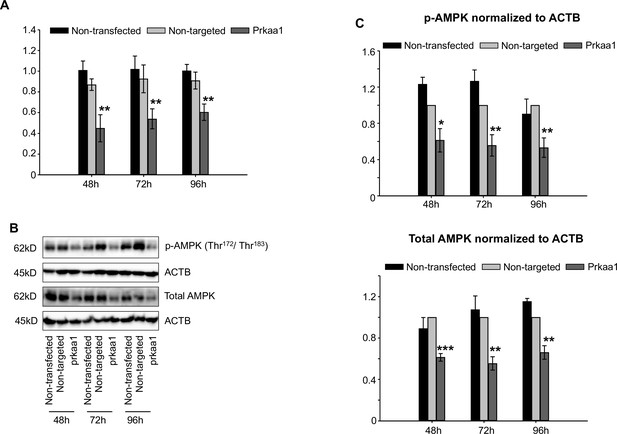
Knockdown of AMP-activated protein kinase (AMPK) by siRNA transfection.
(A) Levels of prkaa1 mRNA in RAW 264.7 cells transfected with prkaa1 or non-targeted control siRNA for 24 hr, followed by incubation for an additional 24, 48, and 72 hr. The expression level of prkaa1 is normalized to that of the housekeeping gene β-actin. (B) Western blot analysis of p-AMPK and total AMPK from cells treated with the different siRNA. A representative blot is shown. (C) Quantified intensities of p-AMPK and total AMPK bands normalized to loading control (β-actin). *: p<0.05, **: p<0.01, ***: p<0.001 using Student’s t-test.
-
Figure 6—figure supplement 2—source data 1
Western blot analysis of p-AMPK and AMPK levels in RAW264.7 cells treated with siRNAs used in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/87458/elife-87458-fig6-figsupp2-data1-v1.pptx
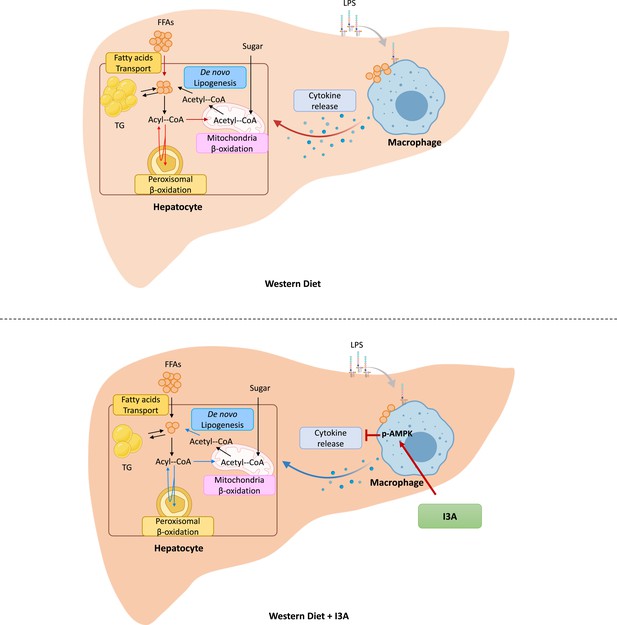
When mice are fed with a Western diet (WD) (top panel), triglycerides (TG) and free fatty acids (FFAs) accumulate in the liver due to increased uptake of fatty acids.
This also leads to increased β-oxidation in the mitochondria and peroxisomes. In liver macrophages, the increase in FFAs, possibly in conjunction with circulating endotoxins (e.g. LPS), stimulates production of inflammatory cytokines. When mice fed the WD are treated with indole-3-acetate (I3A) (bottom panel), both TG and FFAs decrease in the liver. Rather than impact fatty acid uptake, I3A treatment reduces de novo lipogenesis through a downregulation of fatty acid synthase (Fasn), while also reducing both mitochondrial and peroxisomal β-oxidation. In macrophages, I3A attenuates fatty acid and LPS-stimulated production of inflammatory cytokines through activation of AMP-activated protein kinase (AMPK).
Additional files
-
Supplementary file 1
Chromatography gradient method for untargeted LC-MS metabolomics.
Solvent A was formic acid solution in water (0.1% vol/vol). Solvent B was a 0.1% vol/vol formic acid solution in methanol. The flow rate used was 0.4 ml/min.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp1-v1.docx
-
Supplementary file 2
Chromatography gradient and LC-MS parameters used for detection of bile acids.
For the chromatographic method, solvent A was ammonium acetate (2 mM) in water and solvent B was methanol:acetonitrile (50:50, vol/vol). The flow rate used was 0.4 ml/min.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp2-v1.docx
-
Supplementary file 3
Chromatography gradient and LC-MS parameters for free fatty acid analysis.
For the chromatographic method, solvent A was acetonitrile/water (3:2, vol/vol) containing 10 mM ammonium acetate. Solvent B was acetonitrile/isopropanol (1:1, vol/vol). The injection volume was 5 μl and the oven temperature was set to 55°C.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp3-v1.docx
-
Supplementary file 4
Chromatography gradient method used for untargeted proteomics analysis of liver tissue.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp4-v1.docx
-
Supplementary file 5
Data processing workflow used in untargeted proteomic analysis of liver tissue.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp5-v1.pdf
-
Supplementary file 6
LC-MS parameters used for targeted proteomic analysis of liver tissue.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp6-v1.docx
-
Supplementary file 7
Primer sequences used for RT-PCR analysis of gene expression.
- https://cdn.elifesciences.org/articles/87458/elife-87458-supp7-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87458/elife-87458-mdarchecklist1-v1.pdf





