Adulis and the transshipment of baboons during classical antiquity
Figures

Strabo’s reference (17.1.40) to the worship of cynocephali at Hermopolis Magna makes clear that the animal in question is the hamadryas baboon (Papio hamadryas).
The sanctuary and temple complex featured several 35-tonne statues of P. hamadryas as the embodiment of Thoth. One of the oldest deities in the Egyptian pantheon, Thoth is best known as a god of writing and wisdom, a lunar deity, and vizier of the gods, but also as a cosmic deity, creator god, and warrior (Stadler, 2012). The quartzite statues were erected by Amenhotep III, 18th Dynasty, New Kingdom, 1390–1353 BCE. Photograph by N.J. Dominy.
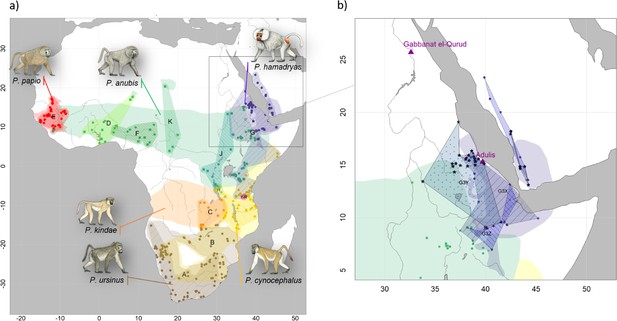
Present-day distributions of the six baboon species, major mitochondrial clades, and provenance of samples analysed in this study.
(a) Overview of species distributions according to the IUCN (2020) and coloured by species (red: P. papio; brown: P. ursinus; yellow: P. cynocephalus; orange: P. kindae; green: P. anubis; purple: P. hamadryas). Colour-patterned regions reflect main mitochondrial clade attribution resulting from phylogenetic reconstructions and are denoted with capital letters A–K (Figure 8). Squares and circles represent geoprovenance of mitogenomes and partial mtDNA datasets (e.g. D-loop, cytochrome b), respectively, and are coloured by species. Note that introgressive hybridization has led to discordances between species assignment and mitochondrial clades. (b) Detailed view of the distribution of mitochondrial subclades G3-X, G3-Y, and G3-Z in the northeastern distribution of baboons. Samples attributed to G3-Y, the subclade assigned to the mummified baboon in phylogenetic reconstructions and haplotype networks, are highlighted with asterisks. The locations of the excavation site of the mummified baboon, Gabbanat el-Qurud, and Adulis are marked with magenta triangles. Male baboon drawings by Stephen Nash, used with permission.
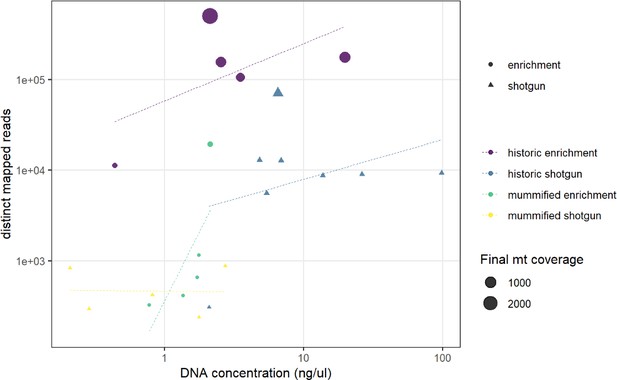
Comparison of DNA concentration and amount of distinct mapped reads.
A higher DNA concentration produces a higher number of distinct mapped reads. Capture enrichment additionally increases the number of distinct mapped reads. Circles and triangles depict the different sequencing approaches, enrichment, and shotgun, respectively; size is related to the final coverage of the mitogenome; colours represent the different sample types and sequencing approaches (yellow: shotgun sequencing of the mummified sample, MHNL 51000172; blue: shotgun sequencing of historic sample; purple: capture enrichment of historic sample; green: capture enrichment of mummy sample).
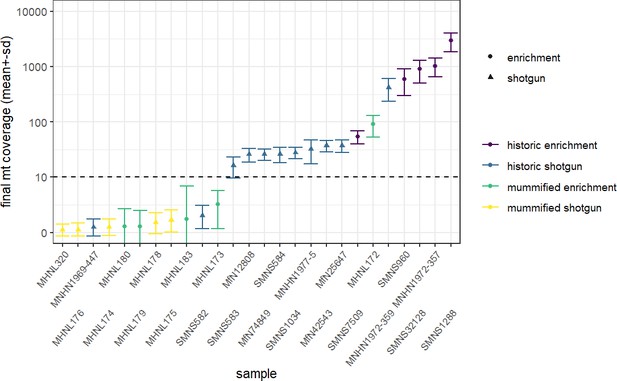
Overview of sequencing success for museum and mummy specimens.
Mean (± SD) final coverage of the mitogenome is shown for each sample (with abbreviated museum ID). Circles and triangles depict the different sequencing approaches, enrichment and shotgun, respectively; colours represent the different sample types and sequencing approaches (yellow: shotgun sequencing of mummy sample; blue: shotgun sequencing of historic sample; purple: capture enrichment of historic sample; green: capture enrichment of mummy sample). Dashed line shows the cut-off limit 10× for mean final coverage; samples below were excluded from final analyses.
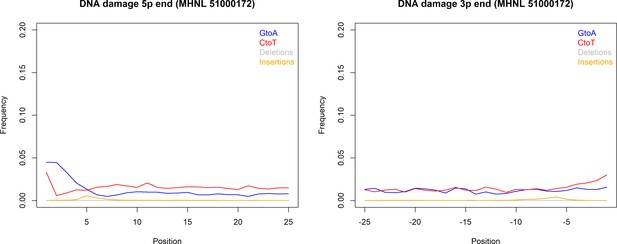
DNA damage plot for the sample of the mummified baboon MHNL 51000172 from 5′ and 3′ read ends, showing mean frequencies of C to T substitutions (red), G to A substitutions (blue), deletions (grey), and insertions (yellow) over the first/last 25 positions.
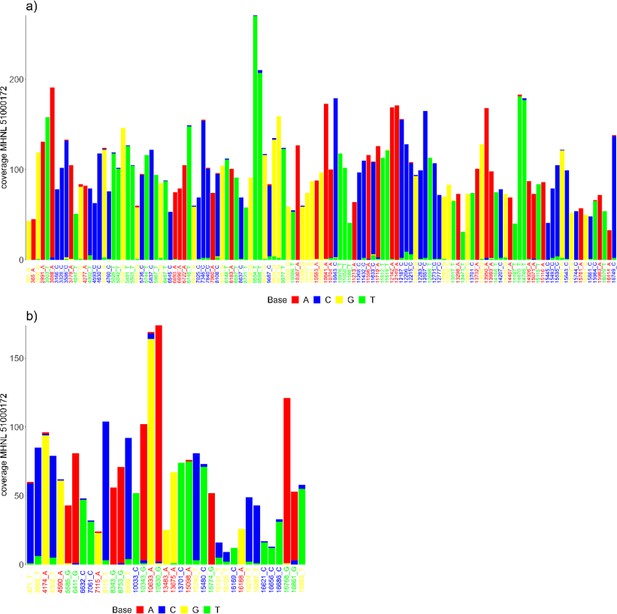
Barplots showing the bases of mapped reads for the sample of the mummified baboon MHNL 51000172 at sites that (a) exhibit fixed differences among northeastern subclades and (b) are fixed in subclade G3-Y but differ in the consensus sequence of the mummified baboon.
Sites are named according to their position and the base in the G3-Y consensus sequence and coloured by base. Bases are colour-coded (A: red; C: blue; G: yellow, T: green).
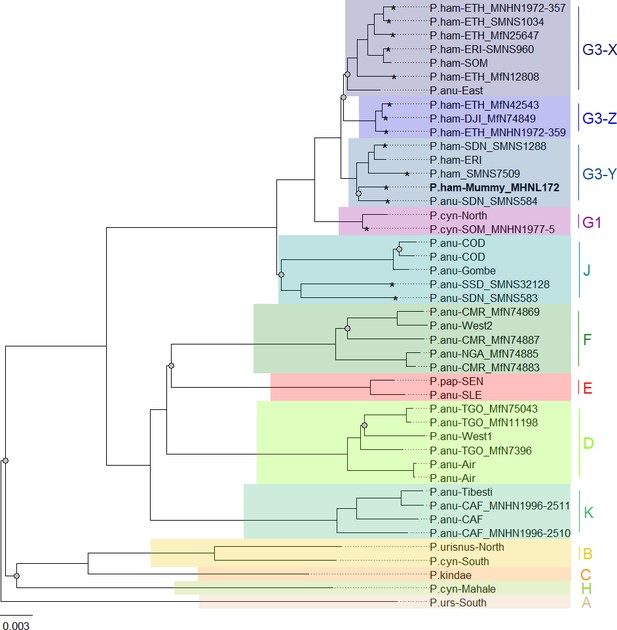
Phylogeny of baboons based on complete mitochondrial genomes as inferred from maximum likelihood analysis.
P. cynocephalus from the Udzungwa Mountains and outgroup T. gelada were omitted from visualization for clarity. The analysed baboon mummy sample MHNL 51000172 (in bold) falls into clade G3-Y. Clade names (A–K) according to Roos et al., 2021, subclades X–Z according to Kopp et al., 2014b; sample IDs include putative species (P.ham, P. hamadryas; P.anu, P. anubis; P.cyn, P. cynocephalus; P.urs, P. ursinus; P.pap, P. papio), country of origin code (CAF, Central African Republic; CMR, Cameroon; COD, Democratic Republic of Congo; DJI, Djibouti; ERI, Eritrea; ETH, Ethiopia; NGA, Nigeria; SDN, Sudan; SSD, South Sudan; SEN, Senegal; SLE, Sierra Leone; SOM, Somalia; TGO, Togo; note that sample SMNS7509 is of unclear geoprovenance), and abbreviated museum code. Nodes with a branch support below 95% are marked with a grey dot. Mitochondrial genomes generated in this study are marked with an asterisk.
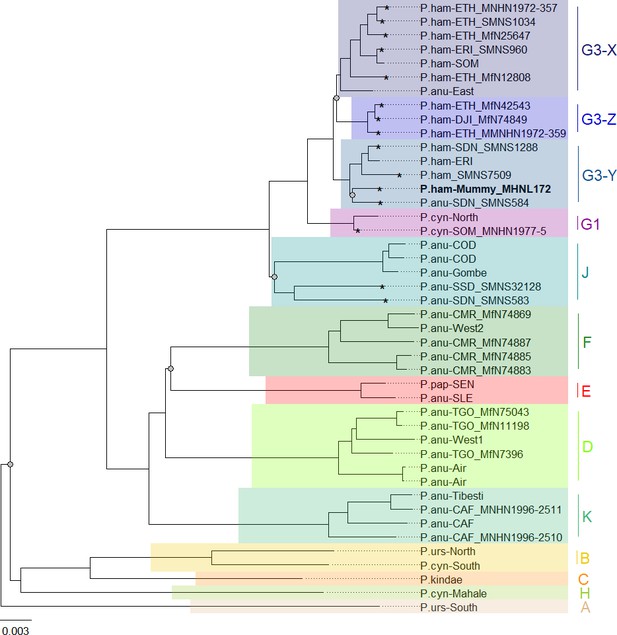
Phylogeny of baboons based on complete mitochondrial genomes under Bayesian inference.
Outgroups (T. gelada and P. cynocephalus-Udzungwa) were omitted from visualization for clarity. The analysed baboon mummy sample MHNL 51000172 (in bold) falls into clade G3-Y. Clade names (A–K) according to Roos et al., 2021, subclades according to Kopp et al., 2014b; sample IDs include putative species (P.ham, P. hamadryas; P.anu, P. anubis; P.cyn, P. cynocephalus; P.urs, P. ursinus; P.pap, P. papio), country of origin code (CAF, Central African Republic; CMR, Cameroon; COD, Democratic Republic of Congo; DJI, Djibouti; ERI, Eritrea; ETH, Ethiopia; NGA, Nigeria; SDN, Sudan; SSD, South Sudan; SEN, Senegal; SLE, Sierra Leone; SOM, Somalia; TGO, Togo; note that sample SMNS7509 is of unclear geoprovenance), and abbreviated museum code. Nodes with PP < 95 are marked with a grey dot. Mitochondrial genomes generated in this study are marked with an asterisk.
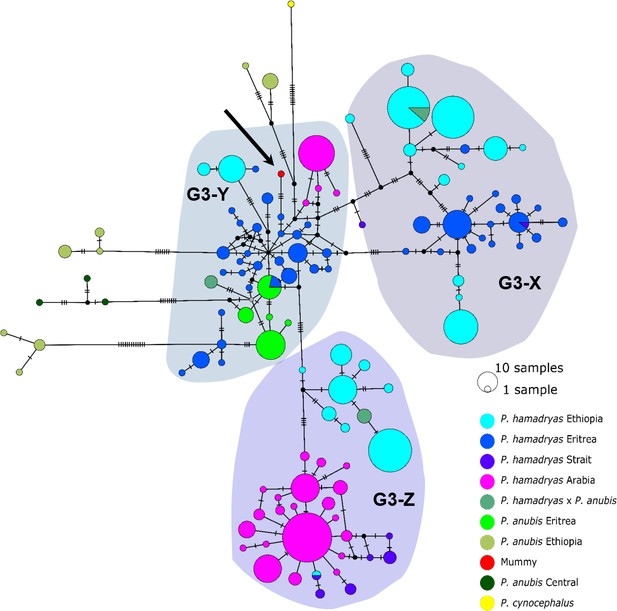
Median-joining haplotype network of northeastern baboons based on 644 HVRI sequences (176 bp).
The analysed baboon mummy sample resolves in clade G3-Y (depicted in red, black arrow). Circle colour reflects species and country of origin (‘Arabia’' comprises samples from Yemen and Saudi Arabia, ‘Strait’ comprises samples from near the Bab-el-Mandab Strait, i.e. southern Eritrea, Djibouti, northern Somalia).
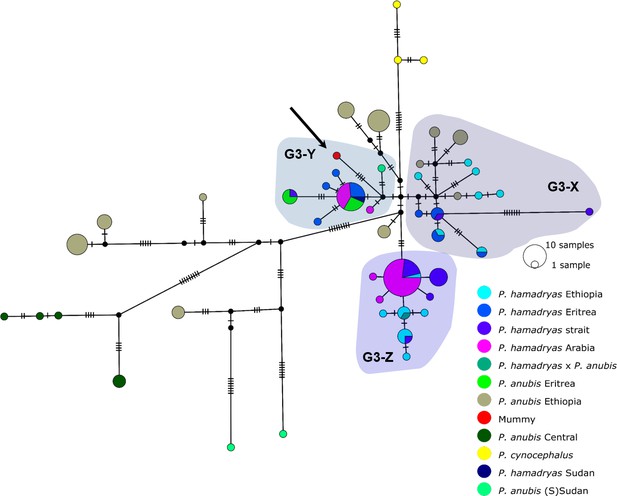
Median-joining haplotype network of northeastern baboons based on 137 cyt b sequences (1140 bp).
The analysed baboon mummy sample resolves in clade G3-Y (depicted in red, arrow). Circle colour reflects species and country of origin (‘Arabia’ comprises samples from Yemen and Saudi Arabia, ‘strait’ comprises samples from near the Bab-el-Mandab Strait, i.e. southern Eritrea, Djibouti, northern Somalia).
Tables
Information on samples analysed in this study.
| Taxon | Origin | Museum ID | Country | Latitude | Longitude | MitoClade | AccNo | Reference |
|---|---|---|---|---|---|---|---|---|
| P. hamadryas | MNHN | MO-1972–357 | ETH | 9.320 | 42.119 | G3-X | OQ538080 | This study |
| P. hamadryas | SMNS | SMNS-Z-MAM-001034* | ETH | 11.500 | 39.300 | G3-X | OQ538076 | This study |
| P. hamadryas | MfN | ZMB_Mam_025647_(2) | ETH | 14.164 | 38.891 | G3-X | OQ538079 | This study |
| P. hamadryas | SMNS | SMNS-Z-MAM-000960 | ERI | 15.783 | 38.453 | G3-X | OQ538078 | This study |
| P. hamadryas | NHMUK | ZD.1910.10.3.1 | SOM | 9.933 | 45.200 | G3-X | MT279063 | Roos et al., 2021 |
| P. hamadryas | MfN | ZMB_Mam_012808 | ETH | 9.314 | 42.118 | G3-X | OQ538089 | this study |
| P. anubis | Wild | ETH | 8.968 | 38.571 | G3-X | JX946196 | Zinner et al., 2013 | |
| P. hamadryas | MfN | ZMB_Mam_042543_(1) | ETH | 9.593 | 41.866 | G3-Z | OQ538084 | this study |
| P. hamadryas | MfN | ZMB_Mam_074849 | DJI | 11.589 | 43.129 | G3-Z | OQ538085 | this study |
| P. hamadryas | MNHN | MO-1972–359 | ETH | 6.998 | 40.478 | G3-Z | OQ538086 | this study |
| P. hamadryas | SMNS | SMNS-Z-MAM-001288 | SDN | 19.110 | 37.327 | G3-Y | OQ538081 | this study |
| P. hamadryas | Wild | ERI | 15.011 | 38.971 | G3-Y | JX946201 | Zinner et al., 2013 | |
| P. hamadryas | SMNS | SMNS-Z-MAM-007509† | - | - | - | G3-Y | OQ538082 | this study |
| P. hamadryas | MHNL | 51000172 | EGY | - | - | G3-Y | OQ538083 | this study |
| P. anubis | SMNS | SMNS-Z-MAM-000584 ‡ | SDN | 13.460 | 33.780 | G3-Y | OQ538075 | this study |
| P. cynocephalus | Wild | TNZ | 7.347 | 37.165 | G1 | JX946199 | Zinner et al., 2013 | |
| P. cynocephalus | MNHN | ZM-MO-1977-5 | SOM | 3.243 | 45.471 | G1 | OQ538088 | this study |
| P. anubis | NHMUK | ZD1929.4.27.2 | COD | 0.800 | 26.633 | J | MT279061 | Roos et al., 2021 |
| P. anubis | NHMUK | ZD1929.4.27.1 | COD | 1.183 | 27.650 | J | MT279062 | Roos et al., 2021 |
| P. anubis | Wild | 19GNM2220916 | TNZ | 4.679 | 29.621 | J | MG787545 | Roos et al., 2018 |
| P. anubis | SMNS | SMNS-Z-MAM-032128 | SSD | 4.281 | 33.555 | J | OQ538087 | this study |
| P. anubis | SMNS | SMNS-Z-MAM-000583 | SDN | 13.333 | 32.729 | J | OQ538077 | this study |
| P. anubis | MfN | ZMB_Mam_074869 | CMR | 5.533 | 12.317 | F | OQ538071 | Kopp et al. in prep |
| P. anubis | Wild | NGA | 7.317 | 11.583 | F | JX946198 | Zinner et al., 2013 | |
| P. anubis | MfN | ZMB_Mam_074887 | CMR | 9.328 | 12.946 | F | OQ538069 | Kopp et al. in prep |
| P. anubis | MfN | ZMB_Mam_074885 | NGA | 7.298 | 10.318 | F | OQ538064 | Kopp et al. in prep |
| P. anubis | MfN | ZMB_Mam_074883 | CMR | 6.334 | 9.961 | F | OQ538072 | Kopp et al. in prep |
| P. papio | Wild | SEN | 12.883 | 12.767 | E | JX946203 | Zinner et al., 2013 | |
| P. anubis | NHMUK | ZD.1947.586 | SLE | 8.917 | 11.817 | E | MT279064 | Roos et al., 2021 |
| P. anubis | MfN | ZMB_Mam_075043 | TGO | 9.260 | 0.781 | D | OQ538066 | Kopp et al. in prep |
| P. anubis | MfN | ZMB_Mam_011198 | TGO | 6.228 | 1.478 | D | OQ538067 | Kopp et al. in prep |
| P. anubis | Wild | CIV | 8.800 | 3.790 | D | JX946197 | Zinner et al., 2013 | |
| P. anubis | MfN | ZMB_Mam_007396_(1) | TGO | 6.950 | 0.585 | D | OQ538065 | Kopp et al. in prep |
| P. anubis | NHMUK | ZD.1939.1022 | NER | 17.000 | 7.933 | D | MT279065 | Roos et al., 2021 |
| P. anubis | NHMUK | ZD.1939.1020 | NER | 17.683 | 8.483 | D | MT279066 | Roos et al., 2021 |
| P. anubis | MNHN | ZM-MO-1960-476 | TCD | 20.344 | 16.786 | K | MT279067 | Roos et al., 2021 |
| P. anubis | MNHN | MO-1996-2511 | CAF | 3.905 | 17.922 | K | OQ538068 | Kopp et al. in prep |
| P. anubis | NHMUK | ZD.1907.7.8.11 | CAF | 8.000 | 20.000 | K | MT279068 | Roos et al., 2021 |
| P. anubis | MNHN | MO-1996-2510 | CAF | 4.966 | 18.701 | K | OQ538070 | Kopp et al. in prep |
| P.ursinus | Wild | ZAF | 24.680 | 30.790 | B | JX946205 | Zinner et al., 2013 | |
| P. cynocephalus | Wild | TNZ | 11.261 | 37.514 | B | JX946200 | Zinner et al., 2013 | |
| P. kindae | ZMB | 12.591 | 30.252 | C | JX946202 | Zinner et al., 2013 | ||
| P. cynocephalus | Wild | 04MNM1300916 | TNZ | 6.119 | 29.730 | H | MT279069 | Roos et al., 2021 |
| P. ursinus | Wild | ZAF | 34.456 | 20.407 | A | JX946204 | Zinner et al., 2013 | |
| P. cynocephalus | Wild | 24UNF1150317 | TNZ | 7.815 | 36.895 | MT279060 | Roos et al., 2021 | |
| Theropithecus gelada | FJ785426 | Hodgson et al., 2009 | ||||||
-
AccNo, GenBank accession number; NHMUK, Natural History Museum, London; MNHN, Muséum National d'Histoire Naturelle, Paris; MfN, Museum für Naturkunde, Berlin; SMNS, State Museum of Natural History Stuttgart; MdC, Musée des Confluences, Lyon
-
*
Mislabelled in museum records as T. gelada.
-
†
Unclear provenance ‘Somaliland’ (not equal to present-day Somaliland).
-
‡
Misidentified provenance ‘Abyssinia’ as Ethiopia in museum records.
Additional files
-
Supplementary file 1
Overview of analysed samples and sequencing results (provided as .csv).
- https://cdn.elifesciences.org/articles/87513/elife-87513-supp1-v2.csv
-
MDAR checklist
- https://cdn.elifesciences.org/articles/87513/elife-87513-mdarchecklist1-v2.pdf