Structure of the connexin-43 gap junction channel in a putative closed state
Figures
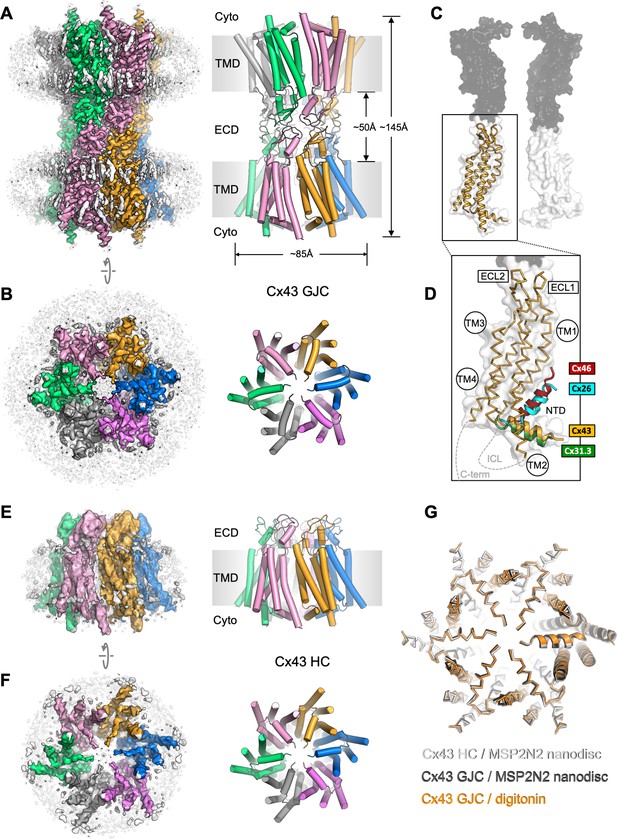
Structure of human connexin-43 (Cx43) gap junction channel (GJC).
(A, B) Cryo-EM density map and model of Cx43 GJC solved by cryo-EM at 2.26 Å resolution. The individual Cx43 monomers in each hemichannel (HC) within the GJC are coloured blue, pink, grey, green, salmon, and orange. Grey densities correspond to the detergent micelle and the bound sterol-like molecules. (C) The position of a single Cx43 monomer (orange) within a GJC (represented as a surface of juxtaposed Cx43 monomers in two distinct membrane regions, white and grey). (D) Alignment of the monomers of Cx43, Cx26 (PDB ID: 2zw3), Cx46 (PDB ID: 7jkc), and Cx31.3 (PDB ID: 6l3t) shows the distinct orientations of the N-terminal domain (NTD) helix. Individual TM domains, extracellular loops 1 and 2 (ECL1–2), relative positions of the intracellular loop (ICL), and the C-terminus (C-term) are indicated. (E, F) Same as A, B, for the Cx43 HC in MSP2N2 nanodiscs at 3.98 Å resolution. (G) Alignment of three indicated structures shows that the conformation of the gating region is highly conserved. The models are shown as ribbons; one of the aligned monomers in each of the structures is represented as cartoon, to clearly indicate the monomer boundaries.
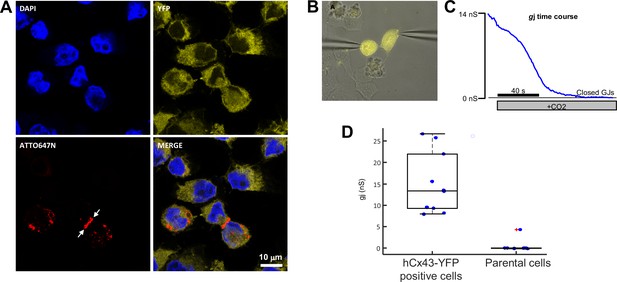
Localization and function of connexin-43 (Cx43) gap junction channels (GJCs).
(A) Immunostaining analysis of transfected HeLa cells expressing Cx43 (red) and cytosolic YFP (yellow). The red fluorescent image of anti-Cx43 ATTO647N-conjugated antibody indicates the presence of a large Cx43 gap junctional region (indicated by white arrows) between neighbouring cells. (B) HeLa cell pair expressing high levels of cytosolic YFP selected to record Cx43 junctional current by dual patch-clamp. (C) gj junctional conductance of the cell pair in B derived in the dual whole-cell configuration. Cell perfusion with saturated CO2 extracellular solution was here used to verify that the junctional current was mediated by connexin channels. (D) Box plot of gj measured in YFP-positive (n = 10) and parental (n = 10) HeLa cell pairs. Mean ± STD ( standard deviation) is 15.2 ± 7.2 and 0.5 ± 1.4 nS, respectively. Statistical analysis was performed using the Mann–Whitney U test, yielding p = 0.0002.
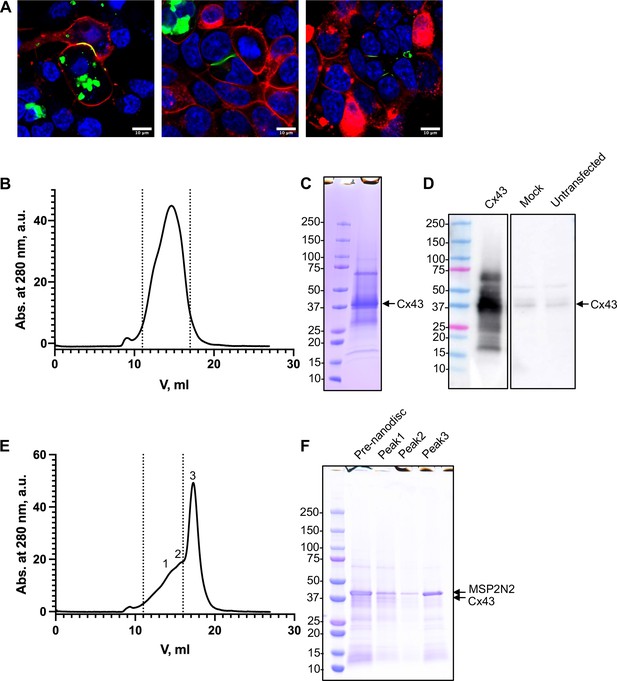
Expression and purification of connexin-43 (Cx43).
(A) Live cell microscopy images of HEK293F cells expressing Cx43; Cx43 was fused with a C-terminal YFP tag (green). The membrane was stained with Vybrant CM-Dil (red) and the nuclei were stained with Hoechst 33342 (blue). The gap junction was formed on the cell membrane between two cells. Size exclusion chromatography (SEC; B) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE; C) of Cx43 purification. The dashed line indicates the fractions of the protein used for the experiments. (D) Western blot analysis of lysates of cells expressing Cx43 (tag-free), using an anti-Cx43 antibody. For comparison, cells mock transfected with an empty plasmid and untransfected HEK293F cells are shown. The major band corresponding to the full-length Cx43 is similar to that in C. (E, F) Same as B, C, for the MSP2N2 nanodisc-reconstituted Cx43 sample. The numbers correspond to the SEC fractions of the protein.
-
Figure 1—figure supplement 2—source data 1
The data corresponding to the panel shown in Figure 1—figure supplement 2B (size exclusion chromatography raw data).
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data1-v1.zip
-
Figure 1—figure supplement 2—source data 2
The data corresponding to the panel shown in Figure 1—figure supplement 2C.
A Coomassie blue-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of connexin-43 (Cx43).
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data2-v1.zip
-
Figure 1—figure supplement 2—source data 3
The data corresponding to the panel shown in Figure 1—figure supplement 2C.
A Coomassie blue-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of connexin-43 (Cx43), with individual lanes labelled.
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data3-v1.zip
-
Figure 1—figure supplement 2—source data 4
The data corresponding to the panel shown in Figure 1—figure supplement 2D.
Western blot of connexin-43 (Cx43)-expressing cell lysates (anti-Cx43 antibody).
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data4-v1.zip
-
Figure 1—figure supplement 2—source data 5
The data corresponding to the panel shown in Figure 1—figure supplement 2D.
Western blot of connexin-43 (Cx43)-expressing cell lysates (anti-Cx43 antibody), with lanes labelled and areas used in Figure 1—figure supplement 2D marked using boxes.
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data5-v1.zip
-
Figure 1—figure supplement 2—source data 6
The data corresponding to the panel shown in Figure 1—figure supplement 2E (size exclusion chromatography raw data).
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data6-v1.zip
-
Figure 1—figure supplement 2—source data 7
The data corresponding to the panel shown in Figure 1—figure supplement 2F.
A Coomassie blue-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of connexin-43 (Cx43) reconstituted in MSP2N2 nanodiscs.
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data7-v1.zip
-
Figure 1—figure supplement 2—source data 8
The data corresponding to the panel shown in Figure 1—figure supplement 2F.
A Coomassie blue-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel of connexin-43 (Cx43) reconstituted in MSP2N2 nanodiscs, with individual lanes labelled.
- https://cdn.elifesciences.org/articles/87616/elife-87616-fig1-figsupp2-data8-v1.zip
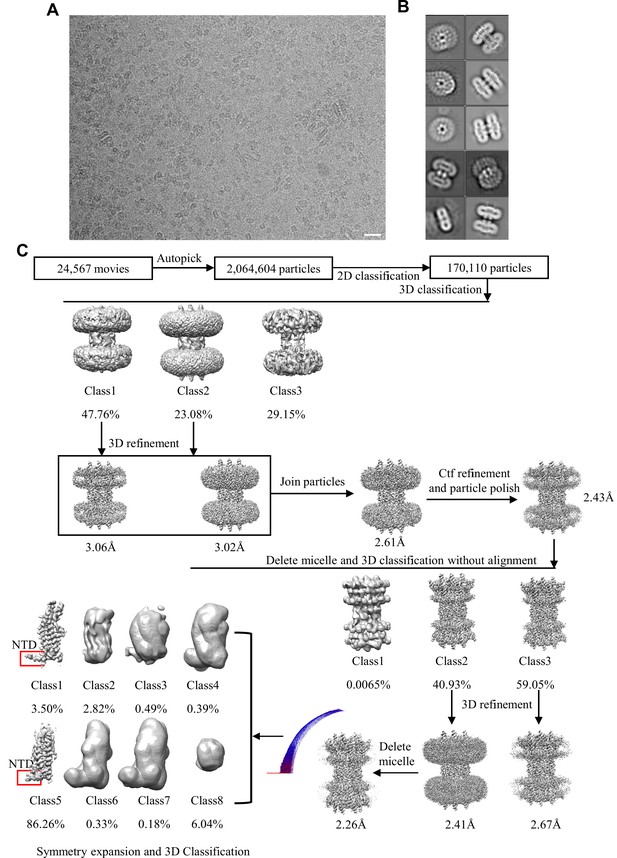
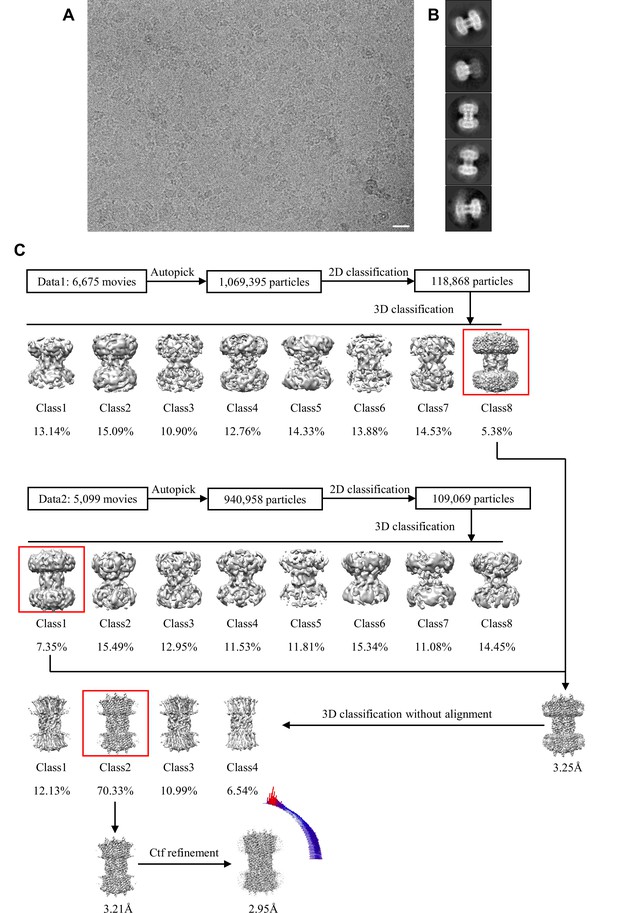
Cryo-EM image processing workflow of connexin-43 (Cx43) in digitonin.
(A) A representative aligned average micrograph of Cx43 sample; the scale bar corresponds to 20 nm. (B) Representative 2D classes of Cx43; the box edge is 260 Å. (C) The overview of the Cx43 cryo-EM image process, including 2D, 3D classification and 3D refinement. Angular distribution histogram of the final reconstruction is represented at the bottom left. Symmetry expansion and 3D classification revealed that the majority of particles cluster into a single class (class 5); a minor class (class 1) showed a similar N-terminal domain (NTD) arrangement.
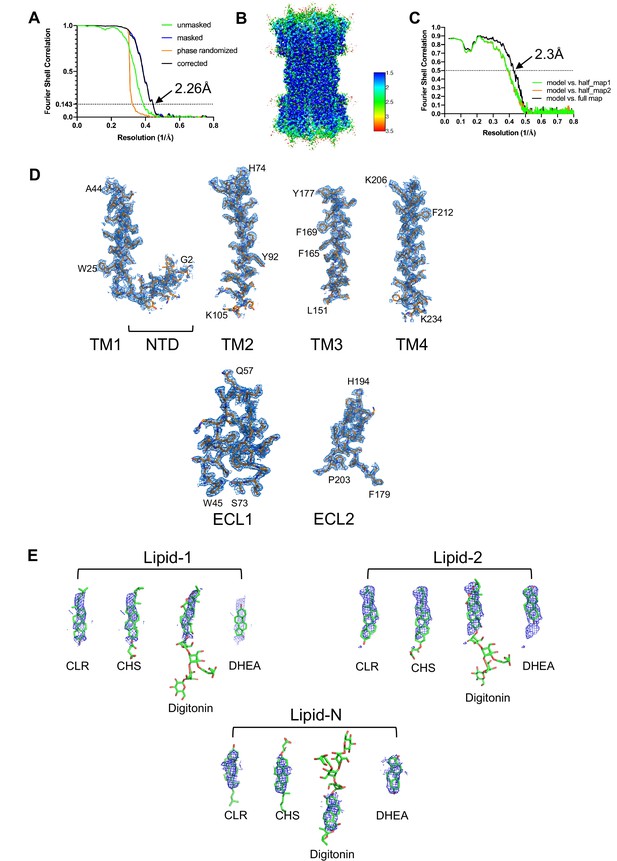
Fourier shell correlation (FSC) of the connexin-43 (Cx43) reconstruction, local resolution and density map features.
(A) Gold-standard FSC plot of Cx43 reconstruction. (B) Local resolution map of Cx43, estimated using ResMap. (C) Model to map FSC plot. (D) Isolated Cx43 density map regions of NTD/TM1, TM2, TM3, TM4, ECL1, and ECL2. (E) A view of the Cx43 lipids densities fit with different possible ligands, CLR, cholesterol hemisuccinate (CHS), Digitonin, or dehydroepiandrosterone (DHEA).
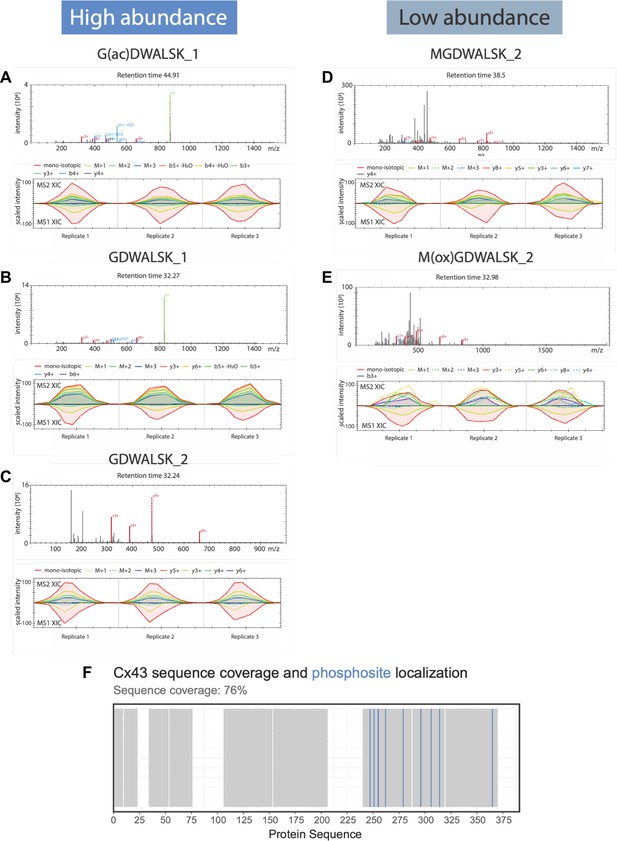
Mass spectrometric characterization of purified connexin-43 (Cx43).
(A–E) Fragment spectra for each of the identified peptides (upper panel) and extracted-ion chromatograms (XIC) for fragment and parent ions detected in MS2 and MS1 spectra (lower panel). (A–C) An N-terminal peptide, G2DWSALGK9, was identified using LC–MS/MS data acquisition and analysis. A fraction of the peptide acetylated at the residue G2 was also observed in the sample, suggesting a mixture of peptide species (C). (D, E) The N-terminal peptides M1GDWSALGK9 and M(ox)1GDWSALGK9 were identified in low abundance, suggesting M1 was deleted during protein expression. (F) Barcode plot showing the identified regions (grey) along the sequence of the purified Cx43 (residues 1–391) and the location of identified phosphosites (blue) with at least 75% confidence at positions 247, 251, 255, 262, 279, 296, 306, 314, 365.
Cryo-EM image processing workflow for connexin-43 (Cx43) gap junction channel (GJC) in nanodisc.
(A) Representative 2D classes of Cx43 GJC in nanodisc sample; the box edge is 260.35 Å. (B-C) The overview of the Cx43 GJC in nanodisc cryo-EM image processing, including 2D (B), 3D classification and 3D refinement (C). Angular distribution histogram of the final reconstruction is represented at the bottom right.
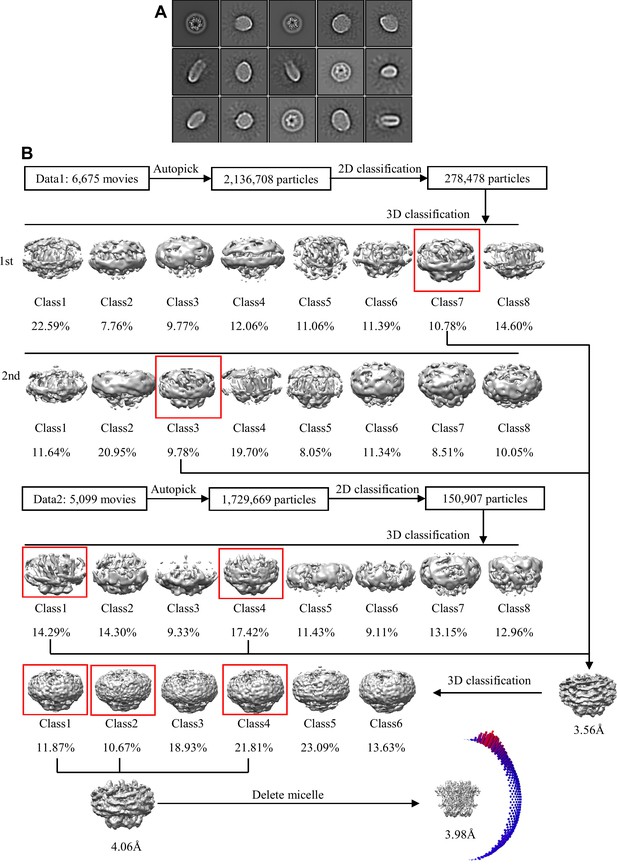
Cryo-EM image processing procedure for connexin-43 (Cx43) hemichannel (HC) in nanodisc.
(A) Representative 2D classes of Cx43 HC in nanodisc sample; the box edge is 260.35 Å. (B) The overview of the Cx43 HC in nanodisc cryo-EM image processing, including 2D, 3D classification and 3D refinement. Angular distribution histogram of the final reconstruction is represented at the bottom right.
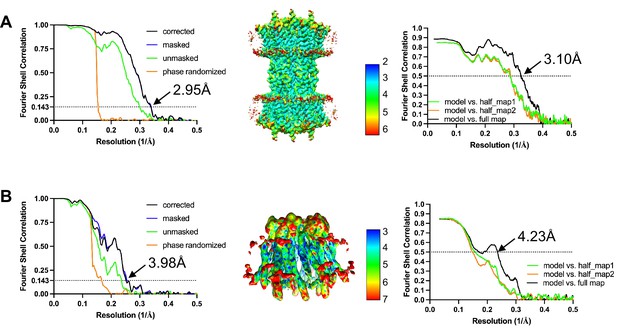
Fourier shell correlation (FSC) and local resolution maps of connexin-43 (Cx43) in nanodiscs.
(A) Gold-standard FSC plot, local resolution, and model to map FSC of Cx43 gap junction channel (GJC) in nanodiscs. (B) Same as A, for Cx43 hemichannel (HC) in nanodiscs.
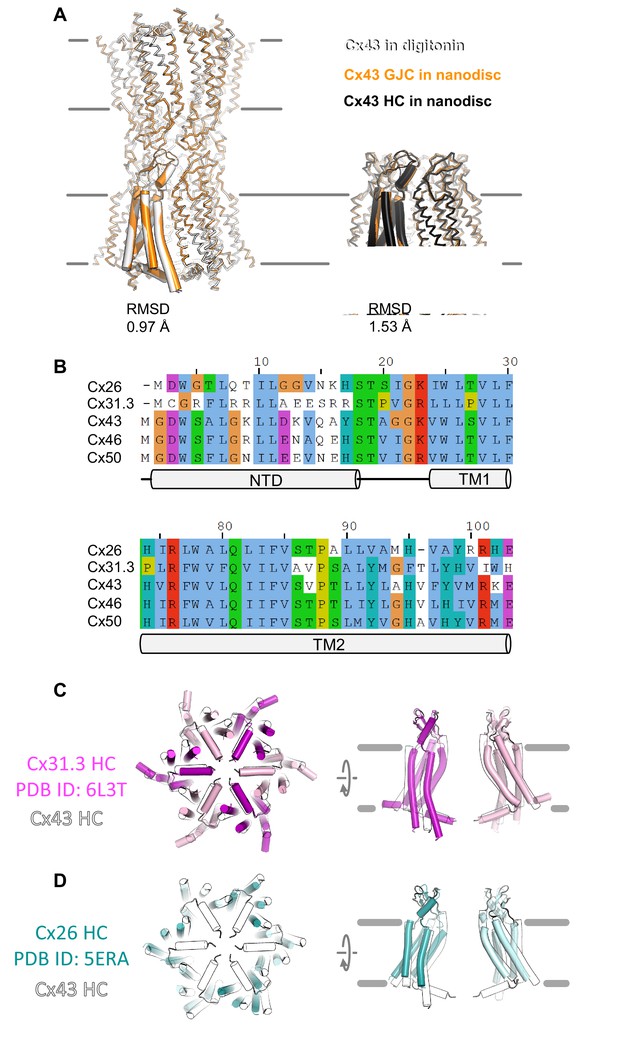
Comparison of the three cryo-EM structures of connexin-43 (Cx43) and sequence alignment of N-terminal domain (NTD) and TM2.
(A) Structural alignment of Cx43 gap junction channels (GJCs) in digitonin and Cx43 in MSP2N2 nanodiscs (left), and for the free Cx43 hemichannels (HCs) and the Cx43 HCs within the MSP2N2 nanodiscs (right). The root-mean-square deviation (RMSD) between the corresponding structures in indicated in the panel. (B) Sequence alignment of the two key structural elements: NTD and TM2, which form the putative cytosolic gating region of Cx43. The sequences used here correspond to the proteins for which structures have been determined experimentally. (C) A comparison of Cx43 HC (white contours) with the Cx31.3 HC structure (pink/magenta; PDB ID: 6L3T). (D) Same as in C, with the Cx26 HC (cyan/teal, used as an HC approximation based on the Cx26 GJC structure; PDB ID: 5ERA).
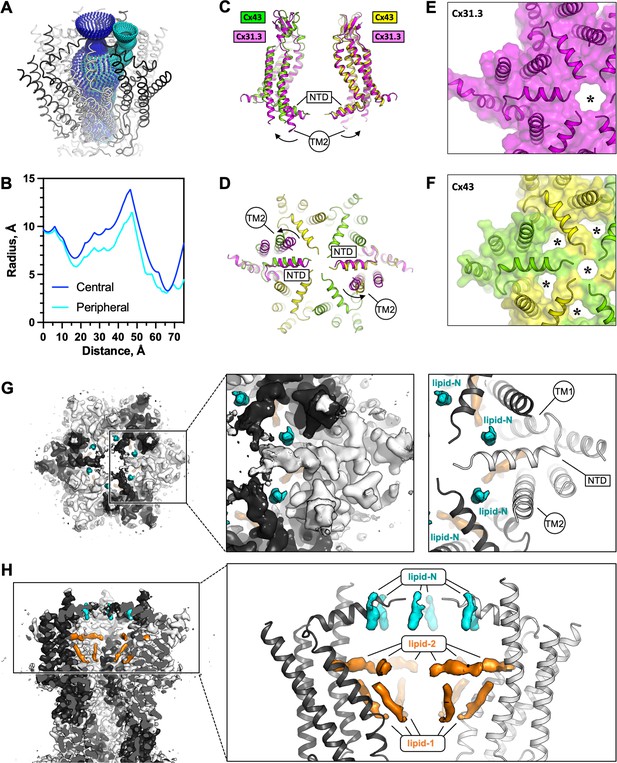
The connexin-43 (Cx43) gate adopts a closed conformation.
(A, B) Analysis of the pore opening dimensions using HOLE reveals a constriction of the pore in the putative gate region. Only a central opening and one of the six peripheral openings within one hemichannel (HC) of Cx43 gap junction channel (GJC) in digitonin is shown. The distance is calculated from the centre of the GJC pore to a point outside of the channel. Central and peripheral openings are coloured blue and cyan, respectively. (C, D) Comparison of the Cx43 in digitonin with the Cx31.3 HC shows that the peripheral opening is created by the particular arrangement of the N-terminal domain (NTD) and by adjustment of the TM2. The distinct NTD/TM2 arrangement results in a single pore opening in the structure of Cx31.3 (indicated with an asterisk, E), contrary to Cx43 (F). (G) A view of the Cx43 gating region from the cytosol reveals the location of the ‘lipid-N’ molecules stabilizing the NTD arrangement, shown as isolated densities (cyan). (H) A slab view of the gating region parallel to the membrane plane shows the relative arrangement of the lipid-N and the intra-pore lipid densities (‘lipid-1’ and ‘lipid-2’; orange).
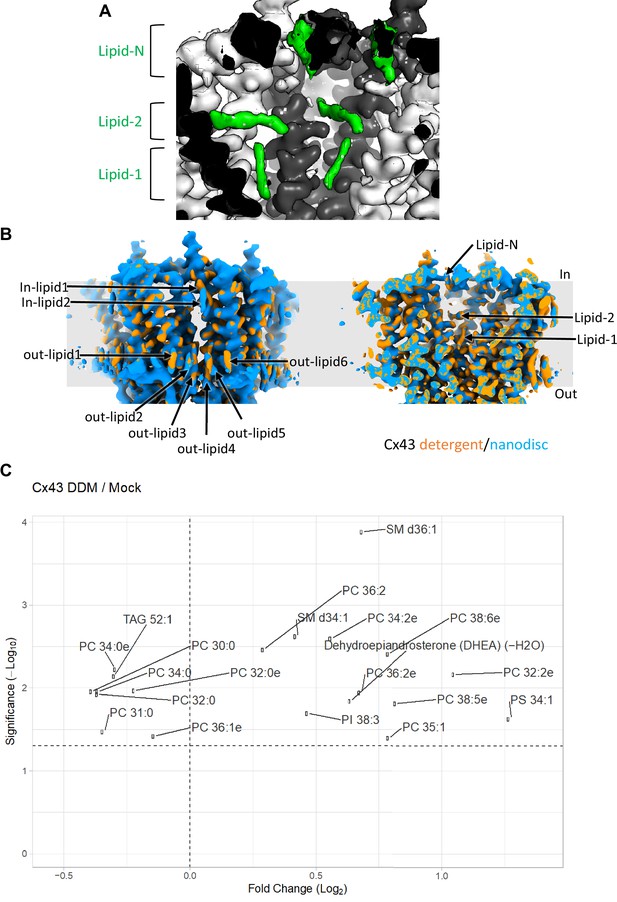
Comparison of intrapore lipid densities in detergent and in nanodiscs and lipidomic analysis of connexin-43 (Cx43).
(A) A view of the intrapore lipid densities in the density map of Cx43 gap junction channel (GJC) in nanodisc; the lipid densities are shown using the unsharpened refined density map, contoured at 4 σ. (B) Comparison of the Cx43 density maps for the detergent and nanodisc samples reveals the conserved positions of the annular lipids (left) and intrapore lipids (right). (C) Lipidomic mass spectroscopy analysis of Cx43 sample versus mock purification. Cx43 was purified in detergent (n-dodecyl-β-D-maltopyranoside, DDM) to avoid introducing any additional sterol molecules into the analysed sample. Mock-purified samples prepared from non-transfected HEK293 cells were used as a mock control. Lipidomic mass spectroscopy analysis was performed to identify lipid molecules co-purified with Cx43.
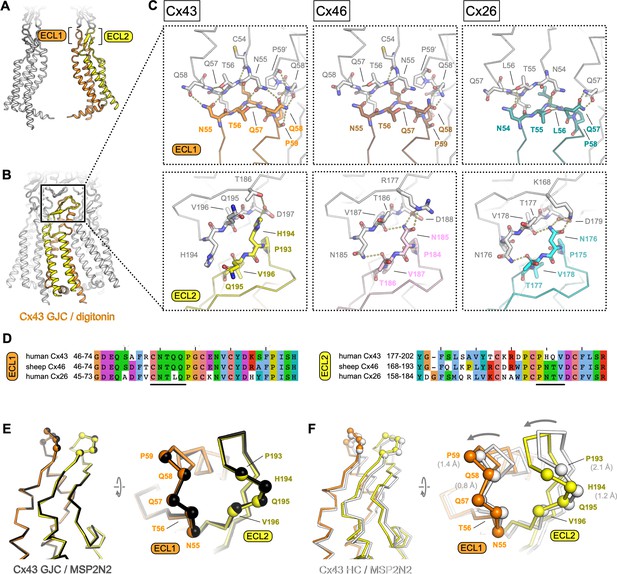
Extracellular domain of connexin-43 (Cx43) gap junction channel (GJC) and hemichannel (HC).
(A, B) The lateral views of Cx43 GJC in digitonin, indicating the positions of the extracellular loops 1 and 2. (C) Views of the extracellular loops ECL1 (top) and ECL2 (bottom), for Cx43 (left), Cx46, (middle; PDB ID: 7jkc), and Cx26 (right; PDB ID: 2zw3). The residues of one monomer in each of the structures, directly involved in junction formation, are coloured by element with carbon atoms as orange/yellow (Cx43), brown/pink (Cx46) and teal/cyan (Cx26). The residues of the neighbouring connexin monomers within 4 Å distance are shown coloured with white carbon atoms. Dotted lines indicate electrostatic contacts, calculated in PyMol. (D) Sequence alignment of the complete ECL1 and ECL2 regions of the three proteins shown in C. The black lines indicate the GJC interface residues, as shown in C. (E) Alignment of the Cx43 GJC structures in digitonin micelles and in nanodiscs. Cα atoms of the interface residues shown in C are represented as spheres (the ribbon and spheres of Cx43 in nanodiscs is coloured black). (F) Same as E, for a comparison between Cx43 GJC in digitonin and Cx43 HC in nanodiscs. The arrow indicates the movement of the two loops that accompanies GJC formation. The ribbon and spheres of Cx43 HC are light grey. The grey numbers in brackets indicate displacement of selected Cα atoms (residues P59, Q58, P193, and P194).
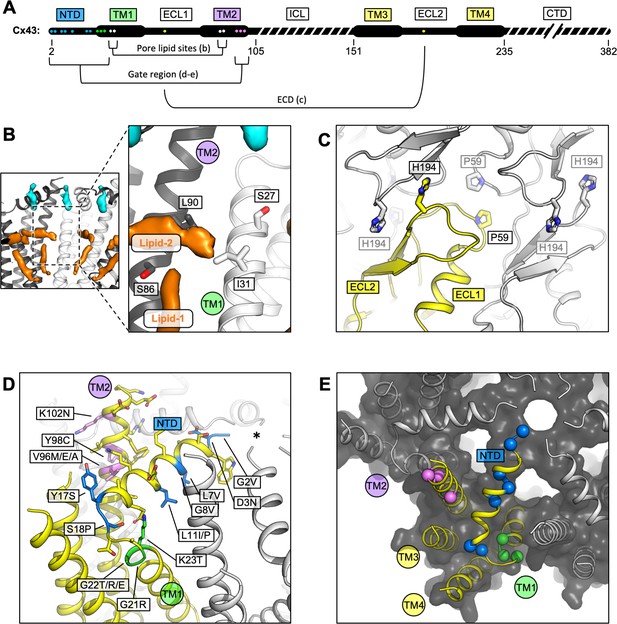
Locations of the disease-linked mutations in the connexin-43 (Cx43) gap junction channel (GJC) structure.
(A) Disease-linked mutations mapped on the sequence of Cx43. The amino acid residues proximal to the intrapore lipid densities (Pore lipid sites) are shown as white dots. The residues at the GJC interface are shown as yellow dots (ECD). The residues of the putative gating region (Gate region) are shown as blue (NTD), green (TM1), and violet (TM2) dots. The sequence elements not resolved in our 3D reconstruction are indicated with a dashed line. (B) Two sterol-like density elements (lipid-1 and -2) are located within the pore region of Cx43 GJC (white). The indicated disease-linked mutations are located within close distance of the pore lipid densities. (C) A view of the extracellular loops forming the junction, indicating two residues known to be linked with oculodentodigital dysplasia (ODDD), P59, and H194 (side chains shown as sticks; one of the monomers in the GJC is coloured yellow). (D) A view of the gating region with the highlighted disease-linked mutations, coloured as in A; the asterisk indicates the center of the gating region. (E) A similar representation of the disease-linked mutations, with Cα atoms shown as spheres, illustrating the contribution of the mutants to gate formation.
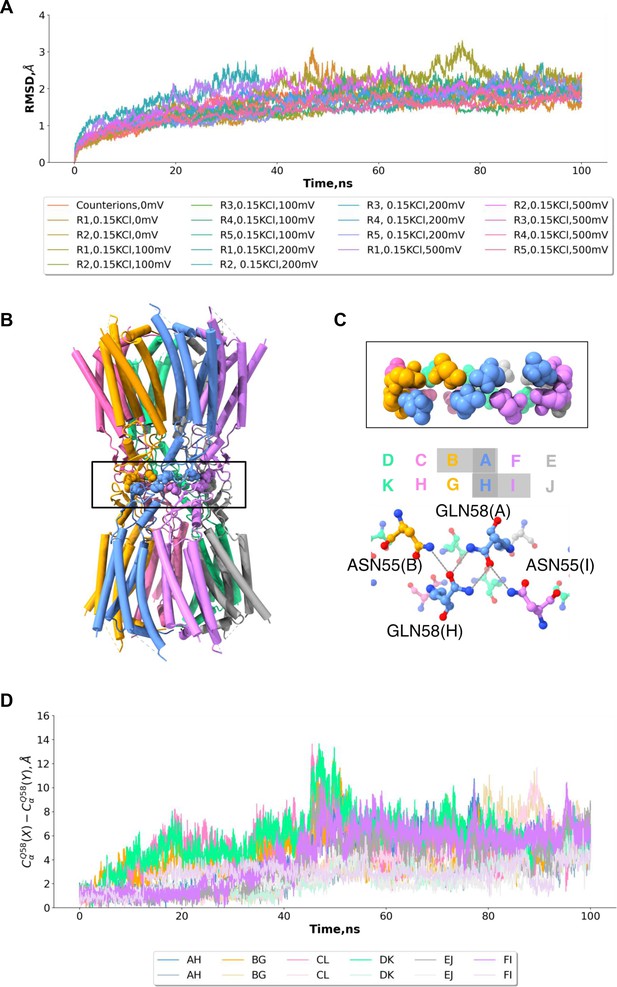
Molecular dynamics simulations of connexin-43 (Cx43) gap junction channel (GJC).
(A) Cα root-mean-square deviation from the cryo-EM structure during the different molecular dynamics simulations. (B) Cx43 GJC structure depicted as cartoon with the interface region boxed. (C) A zoomed-in view of the complex interface highlighted in A and the residues involved in the repeated motif are labelled highlighted as balls and sticks. (D) Distance between the Q58 residues at the interface of the Cx43 during the two simulations at 0 mV and 0.15 KCl.
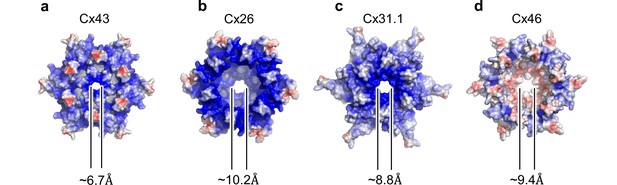
Electrostatic properties and dimensions of the gates in connexin channels.
(a–d) The surface representations of the indicated connexin channels, connexin-43 (Cx43), Cx26 gap junction channel (GJC; PDB ID: 2zw3), Cx31.3 hemichannel (HC; PDB ID: 6l3t), Cx46 GJC (PDB ID: 7jkc), coloured according to the electrostatic potential, calculated using APBS (Smart et al., 1996). The diameters of the gates were calculated using HOLE 71. The images correspond to views of the channel from the cytosol, perpendicular to the membrane plane.
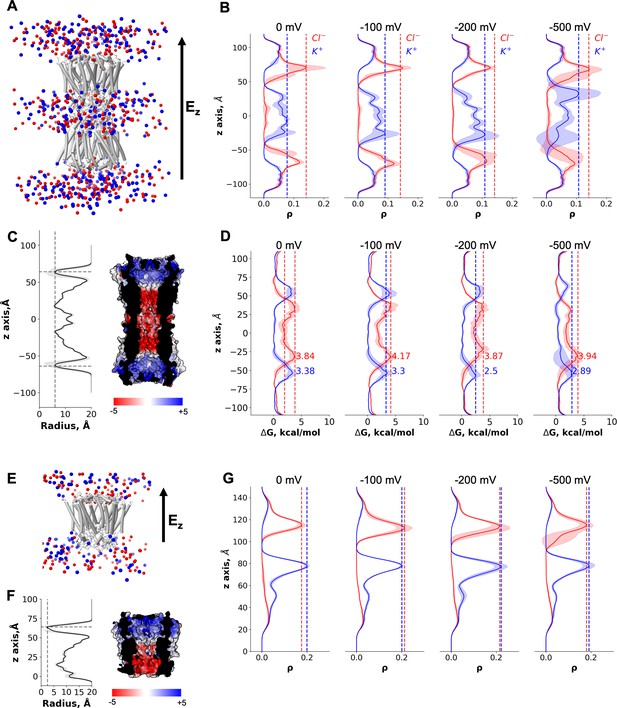
Molecular dynamics simulations of the connexin-43 (Cx43) gap junction channel (GJC) and hemichannel (HC).
(A) Cartoon representation of the double-bilayer system including the GJC (cartoon) and ions (vdw spheres). Lipidic membrane and water residues have been removed for clarity. The direction of the applied constant electric field, Ez, is indicated with an arrow. (B) Ion density (ρ) profiles (average and fluctuations) along the diffusion axis of the GJC coloured in red for anions and blue for cations, for the simulated applied transjunctional voltages. (C) The solvent-accessible radius (and fluctuations during MD) along the diffusion axis of the GJC. The dotted lines correspond to the minimum radius and the position of the N-terminal domain (NTD) regions. The surface representation of Cx43 is coloured according to the calculated electrostatic potential; the slab view shows the properties of the pore. (D) Average free energy experienced (at least two simulation replicates per panel) by the K+ (blue) and Cl− (red) while permeating the pore with different applied transmembrane potential (from right to left: 0, −100, −200, and −500 mV). The dotted lines correspond to the maximum free energy barrier for anions (red) and cations (blue). (E) Cartoon representation of the lipid-bound system including the HC (cartoon) and ions (vdw spheres). Lipidic membrane and water residues have been removed for clarity. The direction of the applied constant electric field, Ez, is indicated with an arrow. (F) Same as C, for the Cx43 HC. (G) Ion density (ρ) profiles (average and fluctuations) along the diffusion axis of the HC coloured in red for anions and blue for cations, for the simulated applied transmembrane voltages.
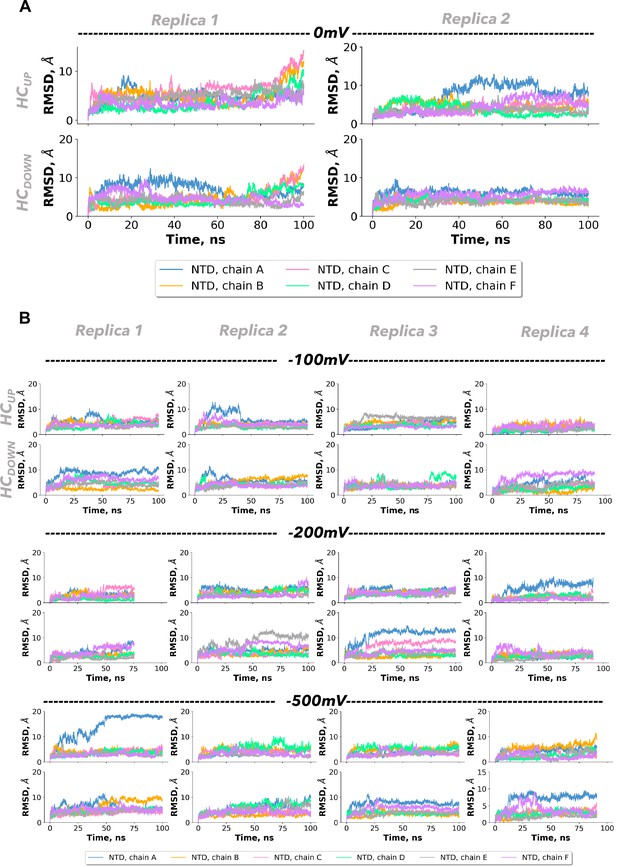
Molecular dynamics simulations of connexin-43 (Cx43) gap junction channel (GJC) at different voltages.
Root-mean-square deviation (RMSD) of the N-terminal domain (NTD) region for the 0 mV replicas (A) and with increasing applied voltage replicas (B). The RMSD from the initial structure over time is depicted for the NTD region of every chain in the dodecameric structure. RMSD is shown for the different simulation replicates from left to right. Each panel includes the data for the chains forming the two hemichannels: for chains A–F (HCUP) and for chains G–L (HCDOWN).
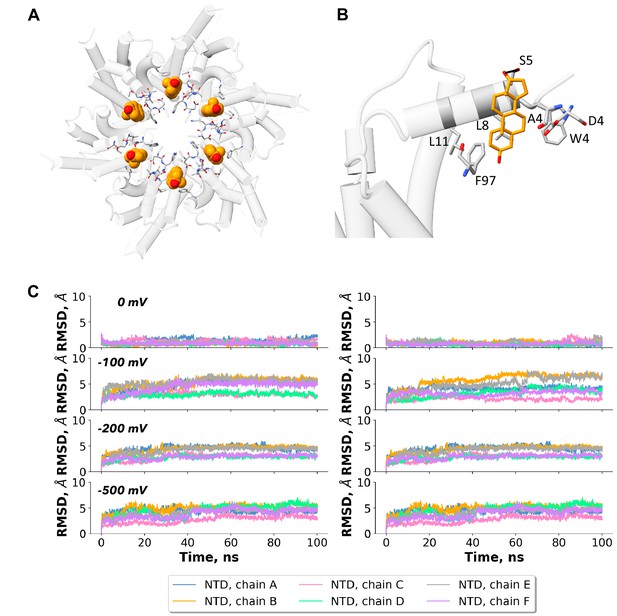
Molecular dynamics (MD) simulations of connexin-43 (Cx43) hemichannel (HC) with dehydroepiandrosterone (DHEA) molecule in lipid-N site.
(A) Cx43 HC structure depicted as a transparent cartoon with the DHEA molecule in orange as Van der Waals spheres and interacting residues from each chain as licorice. The MD simulations were performed with the HC model and a DHEA molecule as a lipid-N surrogate, with the DHEA and the N-terminal domain (NTD) harmonically restrained (as described in Materials and methods). (B) A zoomed-in view of the NTD lipid site, with one DHEA molecule represented as licorice (orange) and the residues involved in contact with the molecule are highlighted as licorice and labelled accordingly. Only one chain is shown as a cartoon for clarity. (C) Root-mean-square deviation (RMSD) of the NTD region for the different replicas (left to right) for the indicated applied voltages (top to bottom).
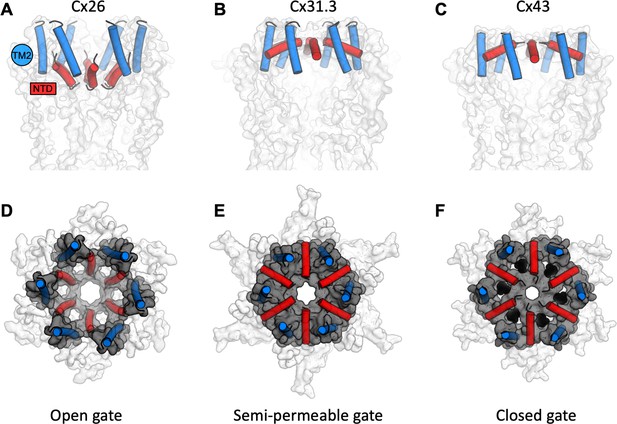
A structure-based view of connexin gating states.
(A) The side view of a fully open gate, as observed in Cx26 gap junction channel (GJC; PDB ID: 2zw3); the gate-forming regions of the protein, N-terminal domain (NTD; red) and TM2 (blue), are shown as cylinders. Similar arrangement of the gating elements has been observed in Cx46/Cx50. This conformation of the gate is permeable to a wide range of substrates, including small molecules and ions. (B) The semi-permeable central gate is featured in the structure of Cx31.3 hemichannel (HC; PDB ID: 6l3t). In this conformation, the gate is likely selective for ions (Cx31.3 has been shown to have a preference for anions), based on the dimensions of the gate. (C) The putative closed state is featured in the connexin-43 (Cx43) structures (here, the Cx43 in digitonin is shown as an example). (D–F) Same as A–C, viewed from the cytosolic side of the channel. The grey surface corresponds to the NTD and TM2 regions; the white surface corresponds to the rest of the protein; lipid-N molecules in F are shown as black spheres, using a model of dehydroepiandrosterone (DHEA) manually placed into the six NTD lipid site regions for illustration purposes (the exact identity of the lipid-N molecule is unknown).
Videos
Molecular dynamics simulation of the connexin-43 (Cx43) gap junction channel (GJC).
N-terminal domain (NTD) of chain A opening upon ion permeation. For clarity only two chains per connexon are depicted as cartoon and coloured white. The corresponding NTDs are coloured according to their conformation position: closed (black), intermediate (‘MD’, red), open (blue). Ions are shown as Van der Waals spheres and coloured according to charge: positive (blue) and negative (red).
Molecular dynamics simulation of the connexin-43 (Cx43) hemichannel (HC), stabilized by dehydroepiandrosterone (DHEA) molecules in the lipid-N sites.
The N-terminal domain (NTD) remains closed during the whole course of the simulation (colours equivalent to those in Video 1).
Tables
Cryo-EM data collection and image processing statistics.
| Data collection | |||
|---|---|---|---|
| Sample | Cx43 (Digitonin) | Cx43_nanodisc | |
| Instrument | FEI Titan Krios/Gatan K3 Summit/Quantum GIF | FEI Titan Krios/Gatan K3 Summit/Quantum GIF | |
| Cx43 GJC | Cx43 HC | ||
| Voltage | 300 | ||
| Electron dose (e−/Å) | 50 | ||
| Defocus range (μm) | −1 to −2 | ||
| Pixel size (Å) | 0.654 | ||
| Map resolution (Å) FSC threshold 0.143 | 2.26 | 2.95 | 3.98 |
| Map sharping b-factor (Å) | −49 | −86 | −100 |
| Number of particles | 50,471 | 10,886 | 20,526 |
| Refinement | |||
| Model resolution (Å) FSC threshold 0.5 | 2.3 | 3.1 | 4.2 |
| Map CC | 0.84 | 0.84 | 0.77 |
| Model composition | |||
| Protein residues | 2268 | 2268 | 1134 |
| ADP (B factor) | 31.90 | 40.02 | 158.95 |
| Bond length RMSD (Å) | 0.003 | 0.003 | 0.003 |
| Bond angle RMSD (°) | 0.552 | 0.514 | 0.678 |
| Validation | |||
| MolProbity score | 1.47 | 1.37 | 1.79 |
| Clash score | 8.80 | 6.76 | 19.75 |
| Rotamer outliers (%) | 0 | 0 | 0 |
| Ramachandran plot | |||
| Favoured (%) | 98.92 | 98.24 | 98.92 |
| Allowed (%) | 1.08 | 1.76 | 1.08 |
| Disallowed (%) | 0 | 0 | 0 |
Ion permeation event statistics.
| Applied transjunctional voltage (mV) | HC1 | HC2 | GJC | |||
|---|---|---|---|---|---|---|
| K+ | Cl− | K+ | Cl− | K+ | Cl− | |
| 0 | 176 ± 30 | 164 ± 56 | 397 ± 119 | 686 ± 122 | 0 | 0 |
| 100 | 275 ± 114 | 126 ± 33 | 490 ± 54 | 803 ± 232 | 1 | 0 |
| 200 | 382 ± 197 | 127 ± 32 | 409 ± 101 | 926 ± 136 | 0 | 0 |
| 500 | 243 ± 149 | 196 ± 115 | 628 ± 86 | 801 ± 270 | 1 | 1 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human) | GJA1 | Uniprot | P17302 | |
| Cell line (human) | Freestyle 293-F cells (HEK293F cells) | Thermo Fischer Scientific; additional authentication not performed | ATCC CRL-1573 | Mycoplasma-negative (fragment length analysis performed by Microsynth) |
| Cell line (human) | HeLa DH cells | Sigma-Aldrich; aitional authentication not performed | ECACC 96112022 | Mycoplasma-negative (fragment length analysis performed by Microsynth) |
| Recombinant DNA reagent | pACMV plasmid | Reeves et al., 2002 | N/A | |
| Recombinant DNA reagent | Homo sapiens Cx43 cDNA | GeneWiz | N/A | Synthetic DNA, cloned into pACMV |
| Peptide, recombinant protein | HRV3C protease | Expressed and purified by Korkhov laboratory | N/A | |
| Commercial assay or kit | NucleoSpin Plasmid | Macherey Nagel | 740588.25 | |
| Commercial assay or kit | NucleoBond Xtra Midi | Macherey Nagel | 740410.5 | |
| Commercial assay or kit | NucleoBond Xtra Maxi Plus | Macherey Nagel | 740416.1 | |
| Chemical compound, drug | Dulbecco’s modified Eagle medium (DMEM) | BioConcept | 1-26F03-I | |
| Chemical compound, drug | Penicillin–streptomycin | PAN Biotech | P06-07100 | |
| Chemical compound, drug | Branched polyethyleneimine (PEI) | Sigma-Aldrich | 408727-100ML | |
| Chemical compound, drug | Trypsin–EDTA PBS 1:250 | BioConcept | 5-51F00-H | |
| Chemical compound, drug | Fetal bovine serum (FBS) | Sigma/Gibco | F0804 | |
| Chemical compound, drug | Benzamidin | Sigma-Aldrich | B6506-5g | |
| Chemical compound, drug | Leupeptin | Sigma-Aldrich | L2884-5mg | |
| Chemical compound, drug | Aprotinin | Sigma-Aldrich | A1153-10mg | |
| Chemical compound, drug | Pepstatin | Sigma-Aldrich | P5318-5mg | |
| Chemical compound, drug | Trypsin inhibitor | Sigma-Aldrich | T9003-100mg | |
| Chemical compound, drug | Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | P7626 | |
| Chemical compound, drug | Deoxyribonuclease I from bovine pancreas (DNase I) | Sigma-Aldrich | DN25-1G | |
| Chemical compound, drug | n-Dodecyl-β-D-maltopyranoside (DDM) | Anatrace | D310 | |
| Chemical compound, drug | Cholesteryl hemisuccinate (CHS) | Sigma-Aldrich | 1510-21-0 | |
| Chemical compound, drug | Digitonin | Merck | 11024-24-1 | |
| Chemical compound, drug | CNBR-activated Sepharose beads | Cytiva | GE17-0430-01 | |
| Chemical compound, drug | Superose 6, 10/300 GL | Cytiva | GE17-5172-01 | |
| Chemical compound, drug | Quantifoil R1.2/1.3 200 mesh Cupper holey carbon grids | PLANO | S143-1 | |
| Chemical compound, drug | Biobeads SM-2 adsorbents | Bio-Rad Laboratories | 152–3920 | |
| Chemical compound, drug | 1-Palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | 850457 C | |
| Chemical compound, drug | Tris–HCl | GERBU | 1028.25 | |
| Chemical compound, drug | Sodium chloride (NaCl) | ROTH | 3957.4 | |
| Chemical compound, drug | 4–20% Mini-PROTEAN TGX Precast Protein Gels | Bio-Rad Laboratories | 4561096DC | |
| Chemical compound, drug | Amicon Ultra-4 Centrifugal Filters Ultracel 100 K | Merck Millipore | UFC810024 | |
| Software, algorithm | Relion | Scheres, 2012. DOI: 10.1016/j.jmb.2011.11.010 | https://www2.mrc-lmb.cam.ac.uk/relion | |
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004. DOI: 10.1002/jcc.20084 | http://www.cgl.ucsf.edu/chimera | |
| Software, algorithm | PHENIX | Adams et al., 2010. DOI:10.1107/S0907444909052925 | https://www.phenix-online.org | |
| Software, algorithm | COOT | Emsley et al., 2010. DOI: 10.1107/S0907444910007493 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | MolProbity | Chen et al., 2010. DOI: 10.1107/S0907444909042073 | http://molprobity.biochem.duke.edu | |
| Software, algorithm | HOLE | Smart et al., 1996. DOI: 10.1016/s0263-7855(97)00,009x | http://www.holeprogram.org/ | |
| Software, algorithm | Pymol | Molecular Graphics System | https://pymol.org/2/ | |
| Software, algorithm | Prism 9.0.0 | GraphPad | https://www.graphpad.com/ | |
| Software, algorithm | GROMACS | Abraham et al., 2015. DOI: https://doi.org/10.1016/j.softx.2015.06.001 | https://www.gromacs.org |





