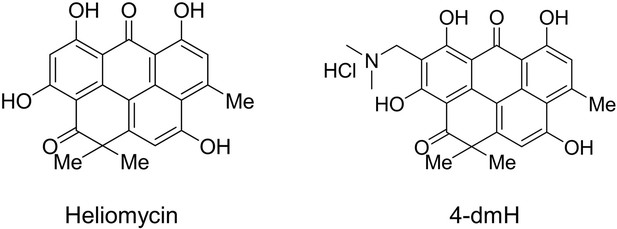
Water-soluble 4-(dimethylaminomethyl)heliomycin exerts greater antitumor effects than parental heliomycin by targeting the tNOX-SIRT1 axis and apoptosis in oral cancer cells
Figures
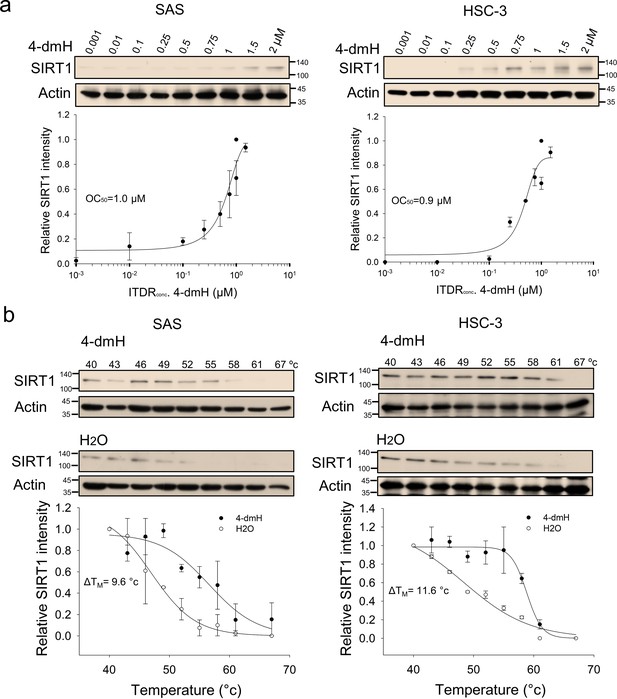
CETSA-based determination of binding between 4-dmH and SIRT1 protein.
(a) Cells were incubated with different concentrations of 4-dmH as described in the Methods. Dose-dependent thermal stabilization of SIRT1 was assessed after heating samples at 54 °C for 3 min in SAS cells and HSC-3 cells. The band intensities of SIRT1 were normalized with respect to the intensity of actin. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) performed in triplicate (n=3). The representative images are generated by SigmaPlot followed by regression wizard. (b) CETSA-melting curves of SIRT1 in the presence and absence of 4-dmH in SAS cells and HSC-3 cells as described in the Methods. The immunoblot intensity was normalized to the intensity of the 40 °C samples. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) performed in triplicate (n=3). The representative images are generated by SigmaPlot followed by regression wizard. The denaturation midpoints were determined using a standard process.
-
Figure 2—source data 1
Original files for the western blot analysis in Figure 2.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig2-data1-v1.zip
-
Figure 2—source data 2
Tiff files containing Figure 2 of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig2-data2-v1.zip
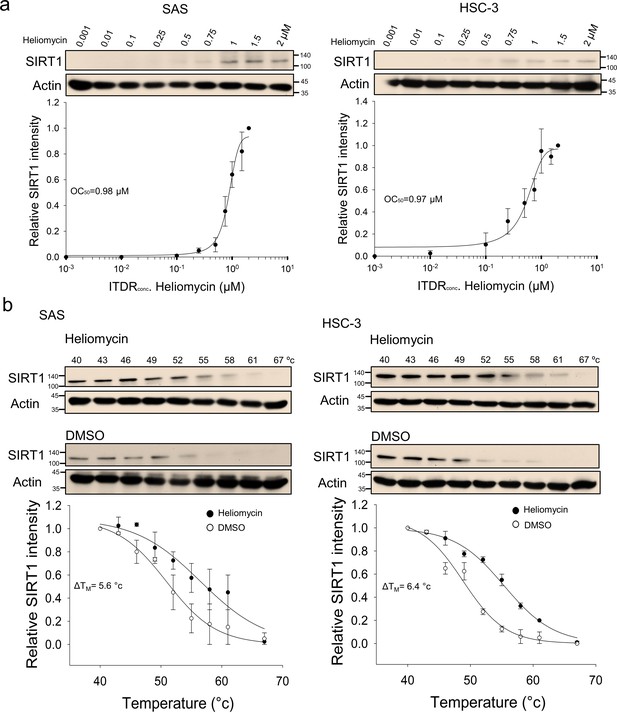
CETSA-based determination of binding between heliomycin and SIRT1 protein.
(a) Cells were incubated with different concentrations of heliomycin as described in the Methods. Dose-dependent thermal stabilization of SIRT1 was assessed after heating samples at 54 °C for 3 min in SAS cells and HSC-3 cells. The band intensities of SIRT1 were normalized with respect to the intensity of actin. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) performed in triplicate (n=3). The representative images are generated by SigmaPlot followed by regression wizard. (b) CETSA-melting curves of SIRT1 in the presence and absence of heliomycin in SAS cells and HSC-3 cells as described in the Methods. The immunoblot intensity was normalized to the intensity of the 40 °C samples. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) performed in triplicate (n=3). The representative images are generated by SigmaPlot followed by regression wizard. The denaturation midpoints were determined using a standard process.
-
Figure 3—source data 1
Original files for the western blot analysis in Figure 3.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig3-data1-v1.zip
-
Figure 3—source data 2
Tiff files containing Figure 3 of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig3-data2-v1.zip
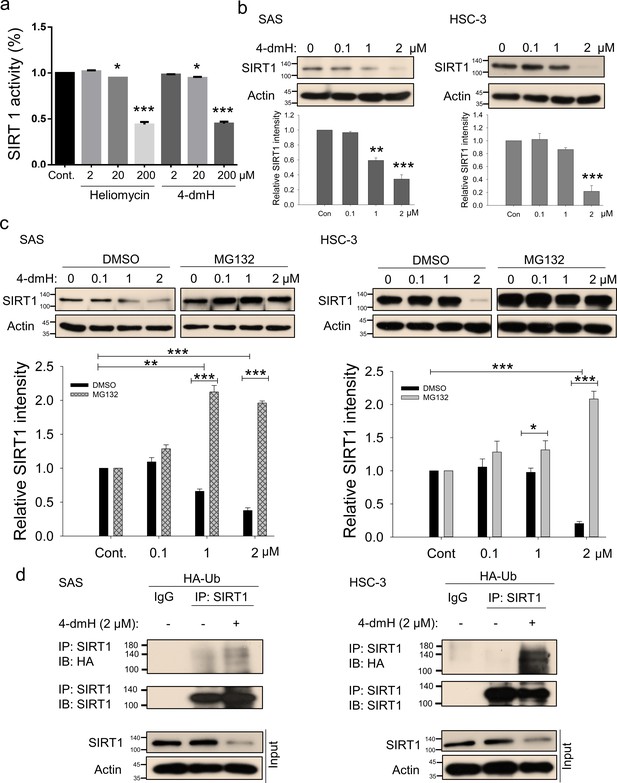
SIRT1 inhibition and downregulation by water-soluble 4-dmH.
(a) SIRT1 activity was determined by a SIRT1 Activity Assay Kit (Fluorometric) with control or recombinant protein treated with test compounds. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as *p<0.05, ***p<0.001. (b) 4-dmH markedly attenuated SIRT1 protein expression analyzed by western blotting. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as **p<0.01, ***p<0.001. (c) 4-dmH-suppressed SIRT1 expression was reverted by MG132, the proteasome inhibitor, in SAS cells and HSC-3 cells. Aliquots of cell lysates were resolved by SDS-PAGE and analyzed by western blotting. β-Actin was detected as an internal control. Representative images are shown. Values (means ± SDs) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as *p<0.05, **p<0.01, *** p<0.001. (d) The lysates of HA-Ub overexpressing cells were immunoprecipitated with nonimmune IgG or an antibody against SIRT1, and the bound proteins were detected by western blotting with anti-HA or anti-SIRT1 antibodies. The total lysates were also immunoblotted with anti-SIRT1 or anti-β-actin antibodies. Aliquots of cell lysates were resolved by SDS-PAGE and analyzed by western blotting. β-actin was detected as an internal control. Representative images are shown.
-
Figure 4—source data 1
Original files for the western blot analysis in Figure 4.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig4-data1-v1.zip
-
Figure 4—source data 2
PDF containing Figure 4b-d of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig4-data2-v1.zip
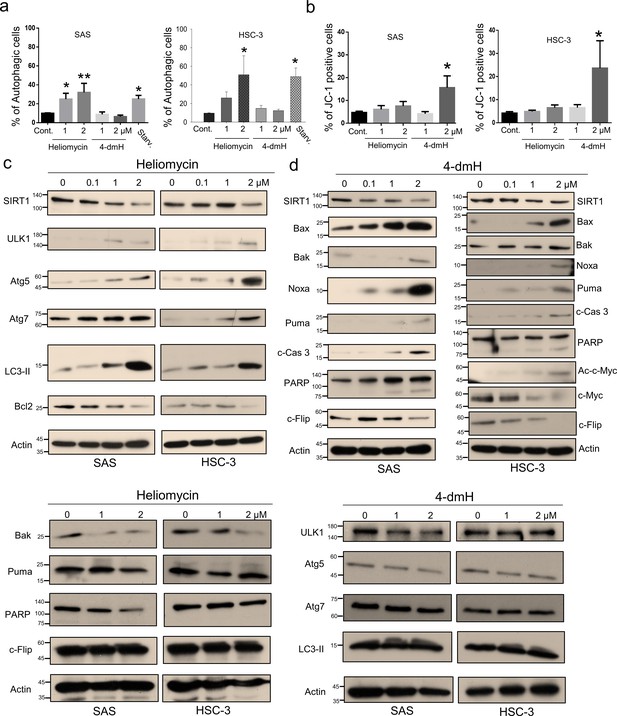
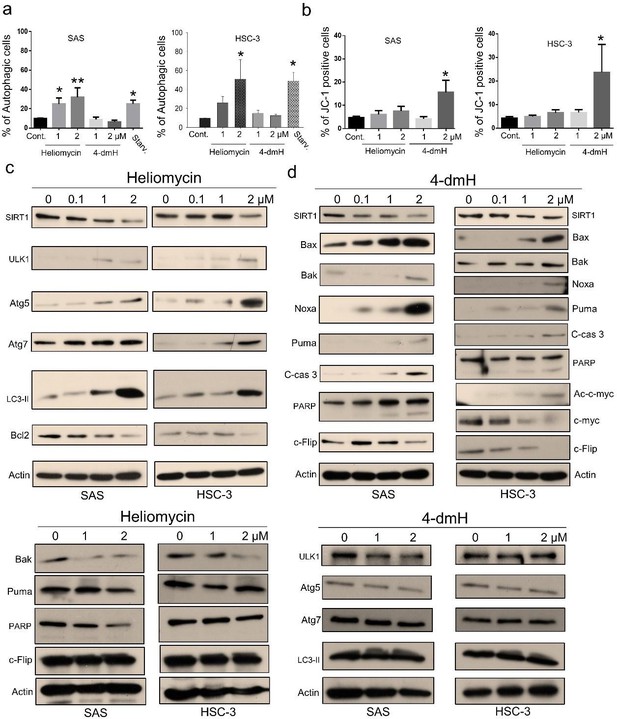
Heliomycin and 4-dmH provoked different cell death pathways in oral cancer cells.
(a) Cells were exposed to heliomycin or 4-dmH and the percentage of the autophagic population was determined by AO staining using flow cytometry. The results are expressed as a percentage of autophagic cells. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as *p<0.05, **p<0.01. (b) Cells were exposed to heliomycin or 4-dmH and the percentage of the apoptotic population was determined by JC-1 staining using flow cytometry. The results are expressed as a percentage of autophagic cells. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant value is as *p<0.05. (c, d) Cells were treated with heliomycin for various concentrations, and aliquots of cell lysates were resolved by SDS-PAGE and analyzed for protein expression by western blotting. β-actin was used as an internal loading control to monitor for equal loading.
-
Figure 5—source data 1
The flow cytometry dot blot autophagy data in Figure 5a.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig5-data1-v1.zip
-
Figure 5—source data 2
The flow cytometry dot blot JC-1 staining data in Figure 5b.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig5-data2-v1.zip
-
Figure 5—source data 3
Original files for the western blot analysis in Figure 5.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig5-data3-v1.zip
-
Figure 5—source data 4
PDF containing Figure 5c,d of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig5-data4-v1.zip
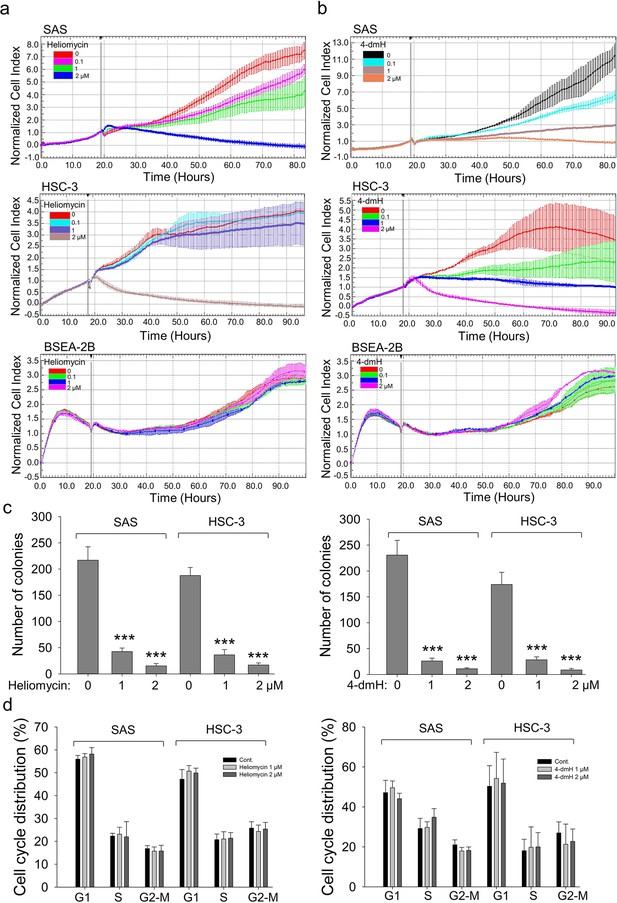
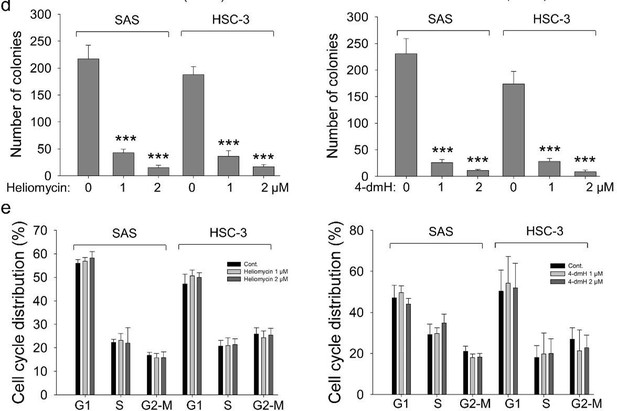
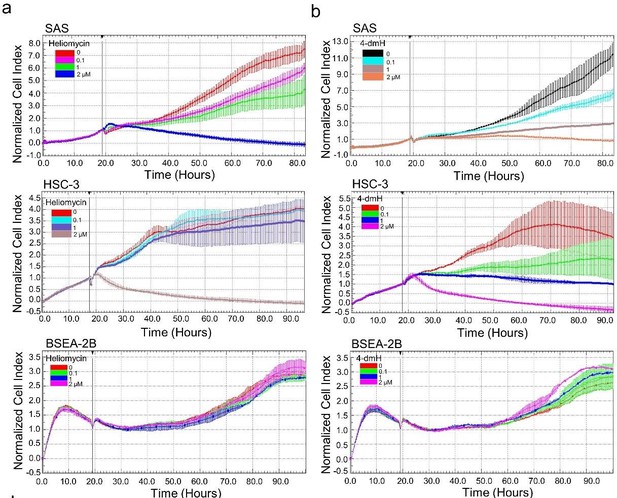
The effects of heliomycin and 4-dmH on proliferation, colony formation, and cell cycle progression of oral cancer cells.
(a, b) Cell proliferation was dynamically monitored by impedance measurements in SAS, HSC-3 cells, and BSEA-2B cells as described in the Methods. Shown are the normalized cell index values measured. (c) Cells were treated with various concentrations of compounds and allowed to form colonies. Colony numbers were determined and documented. Values (means ± SDs) are from three independent experiments (n=3). There was a significant reduction in cells treated with heliomycin or 4-dmH compared to the controls. The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant value is as *** p<0.001. (d) Cells were exposed to different concentrations of compounds for 24 hr and assayed for the cell cycle phase using flow-cytometry. The graphs are representative of three independent experiments (n=3) and generated by SigmaPlot.
-
Figure 6—source data 1
The colony formation representative data in Figure 6c.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig6-data1-v1.zip
-
Figure 6—source data 2
The flow cytometry dot blot cell cycle analysis data in Figure 6d.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig6-data2-v1.zip
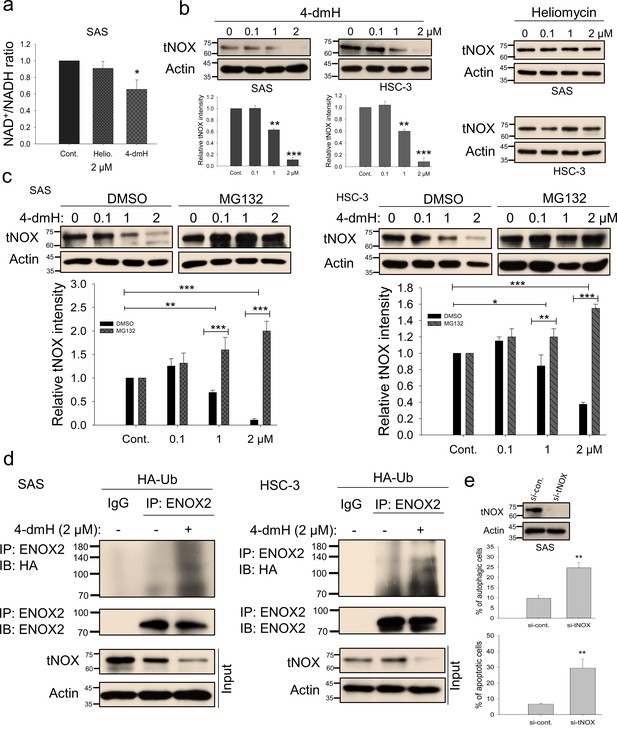
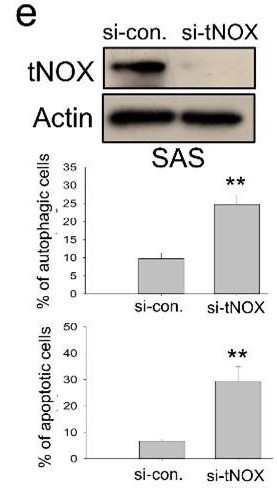
tNOX inhibition and downregulation by water-soluble 4-dmH.
(a) The intracellular NAD+/NADH ratio was measured by an NADH/NAD Quantification Kit (BioVision Inc) with control or lysates of SAS cells treated with heliomycin or 4-dmH. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant value is as *p<0.05. (b) 4-dmH, but not heliomycin, markedly attenuated tNOX protein expression analyzed by western blotting. Values (mean ±SE) are from three independent experiments (n=3) for 4-dmH groups. The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as **p<0.01, *** p<0.001. (c) 4-dmH-suppressed tNOX expression was reversed by MG-132 in SAS cells and HSC-3 cells. Aliquots of cell lysates were resolved by SDS-PAGE and analyzed by western blotting. β-Actin was detected as an internal control. Values (mean ±SE) are from three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant values are as *p<0.05, **p<0.01, *** p<0.001. (d) The lysates of HA-Ub overexpressing cells were immunoprecipitated with nonimmune IgG or an antibody against ENOX2, and the bound proteins were detected by western blotting with anti-HA or anti-ENOX2 antibodies. The total lysates were also immunoblotted with anti-tNOX or anti-β-actin antibodies. (e) The RNA interference-mediated tNOX depletion was conducted in SAS cells for 48 hr. The percentage of apoptotic/autophagic cells was examined by flow cytometry. The presented values (mean ± SDs) represent three independent experiments (n=3). The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant value is as ** p<0.01.
-
Figure 7—source data 1
Original files for the western blot analysis in Figure 7.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig7-data1-v1.zip
-
Figure 7—source data 2
PDF containing Figure 7b-e of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig7-data2-v1.zip
-
Figure 7—source data 3
The flow cytometry dot blot autophagy and apoptosis data in Figure 7e.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig7-data3-v1.zip
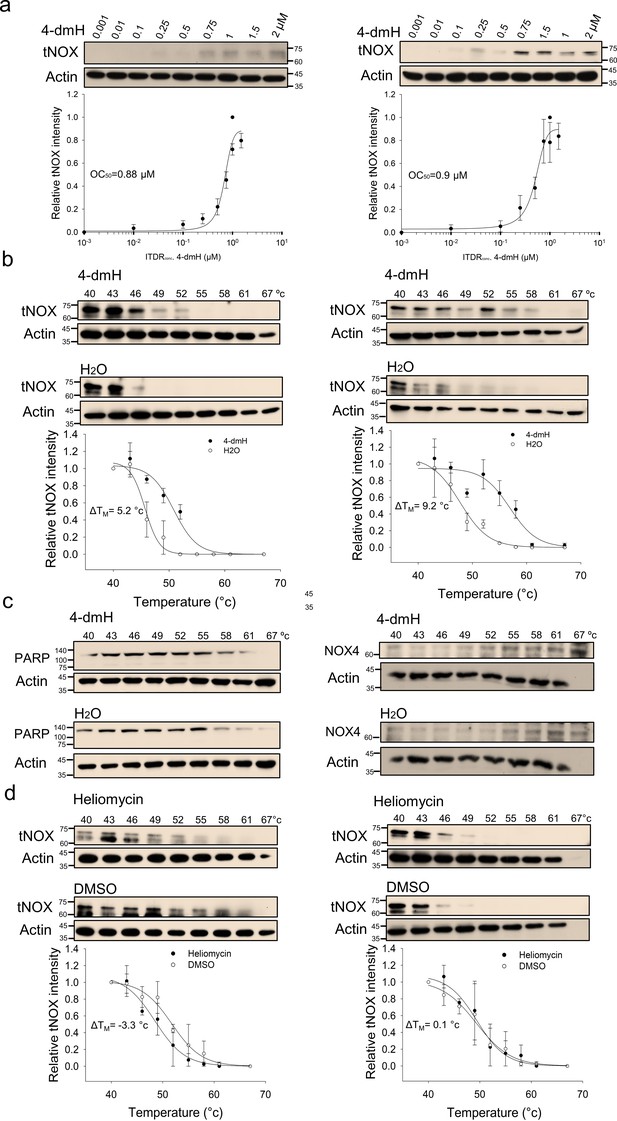
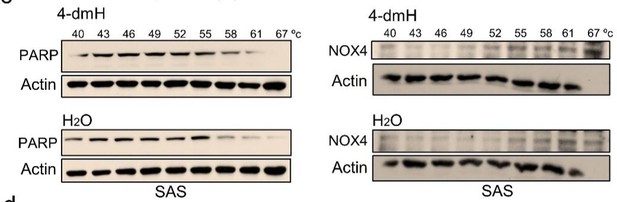
CETSA-based determination of binding between 4-dmH and tNOX protein.
(a) Cells were incubated with different concentrations of the 4-dmH as described in the Methods. Dose-dependent thermal stabilization of tNOX was assessed after heating samples at 54 °C for 3 min in SAS cells and HSC-3 cells. The band intensities of tNOX were normalized with respect to the intensity of actin. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) are from three independent experiments (n=3). The representative images are generated by SigmaPlot followed by regression wizard. (b) CETSA-melting curves of tNOX in the presence and absence of 4-dmH in SAS cells and HSC-3 cells as described in the Methods. The immunoblot intensity was normalized to the intensity of the 40 °C samples. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) are from three independent experiments (n=3). The representative images are generated by SigmaPlot followed by regression wizard. The denaturation midpoints were determined using a standard process. (c) Western blot analysis for PARP or NOX4 in the presence and absence of 4-dmH in SAS cells are shown. The immunoblot intensity was normalized to the intensity of the 40 °C samples. (d) CETSA-melting curves of tNOX in the presence and absence of heliomycin in SAS cells and HSC-3 cells as described in the Methods. The immunoblot intensity was normalized to the intensity of the 40 °C samples. Representative chemiluminescence data are shown on the top. Data are presented as the average and values (mean ±SE) are from three independent experiments (n=3). The representative images are generated by SigmaPlot followed by regression wizard. The denaturation midpoints were determined using a standard process.
-
Figure 8—source data 1
Original file for the western blot analysis in Figure 8.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig8-data1-v1.zip
-
Figure 8—source data 2
PDF containing Figure 8 of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig8-data2-v1.zip
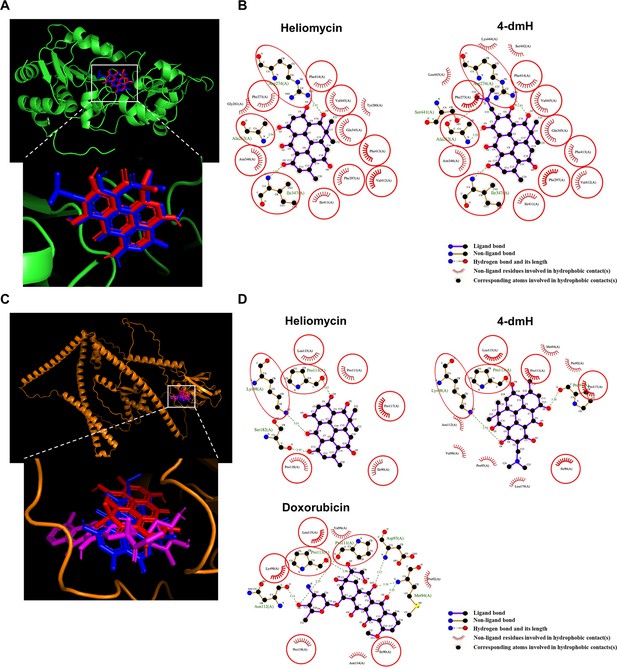
The binding modes of heliomycin and 4-dmH after docking into the pocket of SIRT1 (a, b) and tNOX (c, d).
(a) Superimposition of the docked heliomycin (red) and 4-dmH (blue). (b) Schematic presentations of possible interactions between test compounds and SIRT1 residues. (c) Superimposition of the docked heliomycin (red) and 4-dmH (blue), and doxorubicin (purple). (d) Schematic presentations of possible interactions between test compounds and tNOX residues. The key residues surrounding the binding pocket of SIRT1 and tNOX were identified via the best docking pose of each compound. The red circles and ellipses indicate the identical residues that interacted with different compounds.
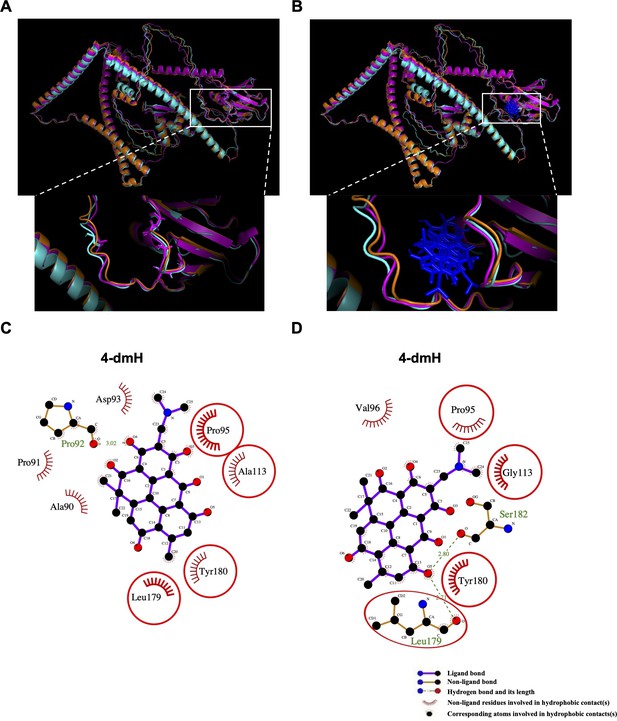
The seven interaction residues on tNOX were substituted with alanine or glycine amino acids and then simulated the protein structures.
The simulated tNOX structures (a, b) and the binding modes of 4-dmH after docking study (c, d). (a) Superimposition of three types of tNOX structures, including the original tNOX structure (orange) and the critical residues in tNOX protein substituted with alanine (magenta) or glycine (cyan). The substituted residues were shown as sticks. (b) Superimposition of the docked 4-dmH (blue). (c) Schematic presentations of possible interactions between 4-dmH and the interacted residues in tNOX protein substituted with alanine. (d) Schematic presentations of possible interactions between 4-dmH and the interacted residues in tNOX protein substituted with glycine. The key residues were identified based on the best docking pose of 4-dmH. The red circles and ellipses indicate the identical residues that interacted with different types of tNOX structures.
-
Figure 9—figure supplement 1—source data 1
The simulated tNOX structures (a, b) and the binding modes of 4-dmH after docking study (c, d).
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig9-figsupp1-data1-v1.zip
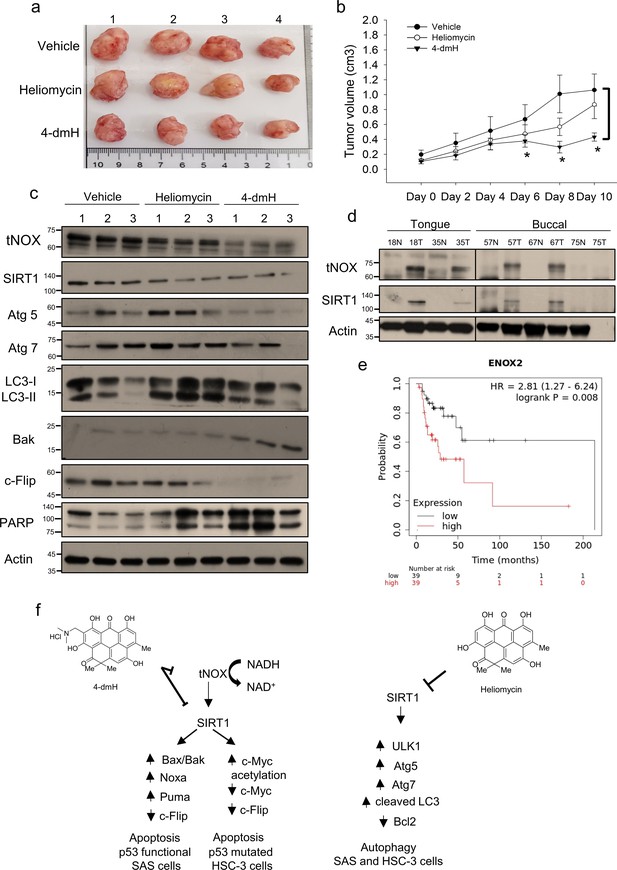
Therapeutic potential of heliomycin and 4-dmH in oral cancer.
(a–c) In a tumor-bearing mouse xenograft model, control mice were intratumorally injected with vehicle buffer and treatment group mice were intratumorally treated with heliomycin or 4-dmH as described in the Methods. The morphology of the tumor tissues excised from tumor-bearing mice (a) and quantitative analysis of xenografted tumor volume during the treatment period (b) are shown. The significance of differences between control and treatment groups was calculated using a one-way ANOVA followed by an appropriate post-hoc test such as LSD. The significant value is as **p<0.05. (c) Tissues from three sets of tumor-bearing mice were grounded and prepared for western blotting analysis. (d) The tumor and adjacent tissues from oral cancer patients were grounded and prepared for western blotting analysis. β-actin was used as an internal loading control to monitor for equal loading. (e) Kaplan-Meier plots of the association between tNOX (ENOX2) expression and overall survival in 78 stage-III head-neck cancer patients. (f) Schematic diagram of the mechanism governing 4-dmH-induced apoptosis and heliomycin-mediated autophagy in oral cancer cells.
-
Figure 10—source data 1
Original files for the western blot analysis in Figure 10.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig10-data1-v1.zip
-
Figure 10—source data 2
PDF containing Figure 10c, d of the relevant western blot analysis with the uncropped gels or blots with the relevant bands.
- https://cdn.elifesciences.org/articles/87873/elife-87873-fig10-data2-v1.zip

The simulated tNOX structures (a, b) and the binding modes of 4-dmH after docking study (c, d).
(a) Superimposition of three types of tNOX structures, including the original tNOX structure (orange) and the critical residues in tNOX protein substituted with alanine (magenta) or glycine (cyan). The substituted residues were shown as sticks. (b) Superimposition of the docked 4-dmH (blue). (c) Schematic presentations of possible interactions between 4-dmH and the interacted residues in tNOX protein substituted with alanine. (d) Schematic presentations of possible interactions between 4-dmH and the interacted residues in tNOX protein substituted with glycine. The key residues were identified based on the best docking pose of 4-dmH. The red circles and ellipses indicate the identical residues that interacted with different types of tNOX structures.